Regulator RcsB
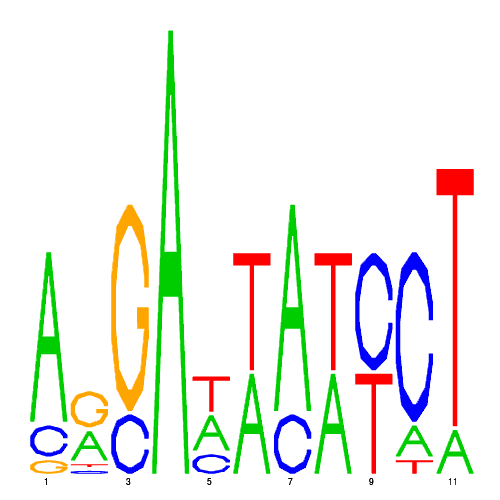
GREs
| GRE ID | PSSM | Score |
|---|---|---|
| eco_265 | 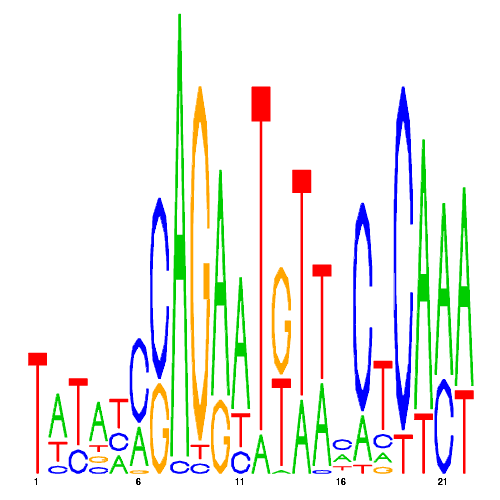 |
0.0018127226850718 |
Corems
| Corem ID | GRE | pval |
|---|---|---|
| ec522616 | eco_265 | 0.0000082160700 |
| ec532853 | eco_265 | 0.0000000000002 |
| ec575306 | eco_265 | 0.0000000000273 |
Genes
| Gene name | Common name | Description | Start | Stop | Strand | Chromosome | GRE | pval |
|---|---|---|---|---|---|---|---|---|
| b0329 | yahO | hypothetical protein | 345708 | 345983 | + | NC_000913.2 | eco_265 | 0.0000000006554 |
| b0419 | yajO | putative NAD(P)H-dependent xylose reductase | 437359 | 436385 | - | NC_000913.2 | eco_265 | 0.0000000289316 |
| b0435 | bolA | possible regulator of murein genes | 453696 | 454013 | + | NC_000913.2 | eco_265 | 0.0000005675291 |
| b0608 | ybdR | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | 641311 | 642549 | + | NC_000913.2 | eco_265 | 0.0000000000864 |
| b0643 | ybeL | hypothetical protein | 674241 | 674723 | + | NC_000913.2 | eco_265 | 0.0000023270920 |
| b0812 | dps | DNA protection during starvation conditions | 848134 | 847631 | - | NC_000913.2 | eco_265 | 0.0000000889639 |
| b0965 | b0965 | orf, hypothetical protein | 1027169 | 1027582 | + | NC_000913.2 | eco_265 | 0.0000012437010 |
| b1259 | yciG | orf, hypothetical protein | 1314059 | 1313880 | - | NC_000913.2 | eco_265 | 0.0000000000000 |
| b1428 | ydcK | predicted enzyme | 1498473 | 1497493 | - | NC_000913.2 | eco_265 | 0.0000039719080 |
| b1480 | sra | 30S ribosomal subunit protein S22 | 1553987 | 1553850 | - | NC_000913.2 | eco_265 | 0.0000000463781 |
| b1536 | ydeI | hypothetical protein | 1622521 | 1622129 | - | NC_000913.2 | eco_265 | 0.0000000232448 |
| b1537 | ydeJ | competence damage-inducible protein A | 1622797 | 1623315 | + | NC_000913.2 | eco_265 | 0.0000000016947 |
| b1724 | ydiZ | hypothetical protein | 1805424 | 1805714 | + | NC_000913.2 | eco_265 | 0.0000033481520 |
| b1810 | yoaC | predicted protein | 1892157 | 1892456 | + | NC_000913.2 | eco_265 | 0.0000000012139 |
| b1955 | yedP | hypothetical protein | 2023535 | 2024350 | + | NC_000913.2 | eco_265 | 0.0000000000000 |
| b2097 | b2097 | orf, hypothetical protein | 2176586 | 2175534 | - | NC_000913.2 | eco_265 | 0.0000000000854 |
| b2112 | yehE | hypothetical protein | 2190818 | 2190537 | - | NC_000913.2 | eco_265 | 0.0000029088650 |
| b3003 | yghA | oxidoreductase | 3147684 | 3148568 | + | NC_000913.2 | eco_265 | 0.0000000554476 |
| b3012 | yqhE | orf, hypothetical protein | 3154645 | 3155472 | + | NC_000913.2 | eco_265 | 0.0000086567490 |
| b4045 | yjbJ | predicted stress response protein | 4257260 | 4257469 | + | NC_000913.2 | eco_265 | 0.0000316466500 |
| b4127 | yjdJ | predicted acyltransferase with acyl-CoA N-acyltransferase domain | 4350108 | 4350380 | + | NC_000913.2 | eco_265 | 0.0000000045360 |
| b4149 | blc | outer membrane lipoprotein (lipocalin) | 4375745 | 4375212 | - | NC_000913.2 | eco_265 | 0.0000047785970 |
Conditions
| Condition ID | Condition Name | GRE | pval |
|---|---|---|---|
| eco_107 | GSM137283 | 230 | 0.0349348800000 |
| eco_116 | GSM18256 | 230 | 0.0191814700000 |
| eco_121 | GSM18264 | 230 | 0.0089455660000 |
| eco_124 | GSM18285 | 230 | 0.0000545710100 |
| eco_133 | GSM37075 | 230 | 0.0393812700000 |
| eco_137 | GSM62194 | 230 | 0.0484863700000 |
| eco_141 | GSM63889 | 230 | 0.0421207200000 |
| eco_149 | GSM67647 | 230 | 0.0489619800000 |
| eco_169 | GSM73258 | 230 | 0.0197139900000 |
| eco_193 | GSM82548 | 230 | 0.0037039520000 |
| eco_195 | GSM82603 | 230 | 0.0130591500000 |
| eco_197 | GSM82616 | 230 | 0.0059194550000 |
| eco_2 | 1285 | 230 | 0.0050583750000 |
| eco_20 | Clean_H2O210 | 230 | 0.0426639700000 |
| eco_203 | GSM83069 | 230 | 0.0016106750000 |
| eco_204 | GSM83071 | 230 | 0.0096216740000 |
| eco_213 | GSM98726 | 230 | 0.0480014600000 |
| eco_221 | GSM99108 | 230 | 0.0064226400000 |
| eco_239 | GSM99173 | 230 | 0.0403221300000 |
| eco_244 | GSM99191 | 230 | 0.0123135000000 |
| eco_247 | GSM99203 | 230 | 0.0394247000000 |
| eco_253 | GSM99393 | 230 | 0.0476228400000 |
| eco_254 | GSM99395 | 230 | 0.0030905390000 |
| eco_256 | GSM99401 | 230 | 0.0476935600000 |
| eco_28 | Clean_TGC2 | 230 | 0.0489308400000 |
| eco_285 | EC_LAC_LA_20_C | 230 | 0.0331646100000 |
| eco_30 | cysB | 230 | 0.0127564900000 |
| eco_312 | GSM106504 | 230 | 0.0318289100000 |
| eco_316 | GSM111419 | 230 | 0.0421852700000 |
| eco_319 | GSM116025 | 230 | 0.0126411700000 |
| eco_326 | GSM118623 | 230 | 0.0290603655000 |
| eco_336 | GSM18275 | 230 | 0.0138602100000 |
| eco_34 | EC_GLY_GLY2_20_A | 230 | 0.0358723200000 |
| eco_340 | GSM23467 | 230 | 0.0031508050000 |
| eco_341 | GSM37064 | 230 | 0.0446255300000 |
| eco_355 | GSM73255 | 230 | 0.0396349700000 |
| eco_356 | GSM73260 | 230 | 0.0436083600000 |
| eco_358 | GSM73266 | 230 | 0.0204607400000 |
| eco_359 | GSM73269 | 230 | 0.0035229670000 |
| eco_361 | GSM73281 | 230 | 0.0109277800000 |
| eco_365 | GSM8113 | 230 | 0.0446452400000 |
| eco_37 | EC_GLY_GLYA_44_B | 230 | 0.0457290200000 |
| eco_378 | GSM83066 | 230 | 0.0393179000000 |
| eco_383 | GSM88912 | 230 | 0.0427494900000 |
| eco_405 | GSM99187 | 230 | 0.0174366300000 |
| eco_416 | 1290 | 230 | 0.0120697500000 |
| eco_417 | 14829 | 230 | 0.0067642230000 |
| eco_420 | 1908 | 230 | 0.0130421700000 |
| eco_424 | Clean_ANA2 | 230 | 0.0259496900000 |
| eco_431 | EC_GLY_GLY2_44_C | 230 | 0.0345875400000 |
| eco_445 | GSM101768 | 230 | 0.0086999840000 |
| eco_448 | GSM106752 | 230 | 0.0265661900000 |
| eco_450 | GSM111416 | 230 | 0.0341038300000 |
| eco_454 | GSM118600 | 230 | 0.0412371200000 |
| eco_459 | GSM18237 | 230 | 0.0310180200000 |
| eco_461 | GSM18284 | 230 | 0.0360171900000 |
| eco_463 | GSM30819 | 230 | 0.0430370100000 |
| eco_465 | GSM37068 | 230 | 0.0077169370000 |
| eco_467 | GSM37074 | 230 | 0.0119305100000 |
| eco_473 | GSM65588 | 230 | 0.0259450300000 |
| eco_475 | GSM65623 | 230 | 0.0364779200000 |
| eco_482 | GSM73123 | 230 | 0.0463690800000 |
| eco_487 | GSM73268 | 230 | 0.0274588000000 |
| eco_488 | GSM73279 | 230 | 0.0005581082000 |
| eco_494 | GSM77355 | 230 | 0.0404009800000 |
| eco_496 | GSM82600 | 230 | 0.0103243800000 |
| eco_500 | GSM88913 | 230 | 0.0048156810000 |
| eco_504 | GSM98728 | 230 | 0.0444323300000 |
| eco_505 | GSM99083 | 230 | 0.0127892200000 |
| eco_507 | GSM99088 | 230 | 0.0202887700000 |
| eco_510 | GSM99102 | 230 | 0.0118895300000 |
| eco_521 | GSM99199 | 230 | 0.0476242500000 |
| eco_530 | 14830 | 230 | 0.0176429200000 |
| eco_535 | Clean_DIAUX5 | 230 | 0.0424544600000 |
| eco_539 | crpcysB__G | 230 | 0.0185005200000 |
| eco_540 | crpfruR__G | 230 | 0.0356016000000 |
| eco_544 | EC_GLY_GLY2_44_A | 230 | 0.0344526900000 |
| eco_561 | E-MEXP-267-raw-data-495080319.txt | 230 | 0.0348343700000 |
| eco_57 | EC_LAC_WILD_E | 230 | 0.0365517900000 |
| eco_576 | GSM18261 | 230 | 0.0337026000000 |
| eco_577 | GSM18287 | 230 | 0.0196641400000 |
| eco_585 | GSM73132 | 230 | 0.0161358100000 |
| eco_596 | GSM83076 | 230 | 0.0108403700000 |
| eco_604 | GSM99123 | 230 | 0.0197185700000 |
| eco_612 | GSM99206 | 230 | 0.0070094550000 |
| eco_614 | GSM99210 | 230 | 0.0000532207200 |
| eco_615 | GSM99399 | 230 | 0.0198423100000 |
| eco_629 | EC_GLY_GLYC_20_A | 230 | 0.0409177400000 |
| eco_63 | E-MEXP-244-raw-data-396455082.txt | 230 | 0.0040985430000 |
| eco_633 | EC_GLY_WILD_E | 230 | 0.0386425400000 |
| eco_634 | EC_LAC_L2_20_A | 230 | 0.0408668700000 |
| eco_64 | E-MEXP-245-raw-data-396455333.txt | 230 | 0.0407134600000 |
| eco_646 | GSM116027 | 230 | 0.0106912800000 |
| eco_676 | GSM73275 | 230 | 0.0019908430000 |
| eco_677 | GSM73280 | 230 | 0.0294059200000 |
| eco_685 | GSM83068 | 230 | 0.0021743910000 |
| eco_686 | GSM83072 | 230 | 0.0263638100000 |
| eco_69 | fur__G | 230 | 0.0098429330000 |
| eco_705 | GSM99397 | 230 | 0.0000942225500 |
| eco_706 | 1912 | 230 | 0.0024523390000 |
| eco_709 | Clean_H2O29 | 230 | 0.0335270100000 |
| eco_724 | GSM35419 | 230 | 0.0108036000000 |
| eco_726 | GSM63888 | 230 | 0.0395245900000 |
| eco_727 | GSM65589 | 230 | 0.0099827250000 |
| eco_734 | GSM73252 | 230 | 0.0027644010000 |
| eco_735 | GSM73278 | 230 | 0.0280273500000 |
| eco_743 | GSM98722 | 230 | 0.0451938300000 |
| eco_744 | GSM99085 | 230 | 0.0048006350000 |
| eco_754 | Clean_ANA17 | 230 | 0.0177951200000 |
| eco_760 | EC_LAC_LD_60_B | 230 | 0.0424433100000 |
| eco_771 | GSM73127 | 230 | 0.0254718600000 |
| eco_775 | GSM73289 | 230 | 0.0002969868000 |
| eco_777 | GSM83059 | 230 | 0.0081465430000 |
| eco_79 | GSM101771 | 230 | 0.0040269330000 |
| eco_793 | GSM116029 | 230 | 0.0426201700000 |
| eco_80 | GSM101772 | 230 | 0.0050505280000 |
| eco_808 | GSM99638 | 230 | 0.0427272700000 |
| eco_81 | GSM101773 | 230 | 0.0143925700000 |
| eco_814 | EC_GLY_WILD_A | 230 | 0.0019904150000 |
| eco_815 | GSM101770 | 230 | 0.0092060400000 |
| eco_817 | GSM18277 | 230 | 0.0443563000000 |
| eco_823 | EC_GLY_GLYB_44_B | 230 | 0.0176094600000 |
| eco_826 | GSM106756 | 230 | 0.0481257000000 |
| eco_829 | GSM67663 | 230 | 0.0438115200000 |
| eco_830 | GSM99106 | 230 | 0.0273605600000 |
| eco_841 | GSM37076 | 230 | 0.0210519400000 |
| eco_844 | GSM99214 | 230 | 0.0360348300000 |
| eco_846 | GSM18248 | 230 | 0.0374506100000 |
| eco_849 | GSM120197 | 230 | 0.0385365900000 |
| eco_850 | GSM88918 | 230 | 0.0283689300000 |
| eco_851 | GSM99112 | 230 | 0.0439770700000 |
| eco_861 | GSM99124 | 230 | 0.0405339900000 |
| eco_863 | GSM73118 | 230 | 0.0074530060000 |
| eco_867 | GSM83075 | 230 | 0.0340815700000 |
| eco_96 | GSM116026 | 230 | 0.0231440000000 |