eco_16209 Co-expression
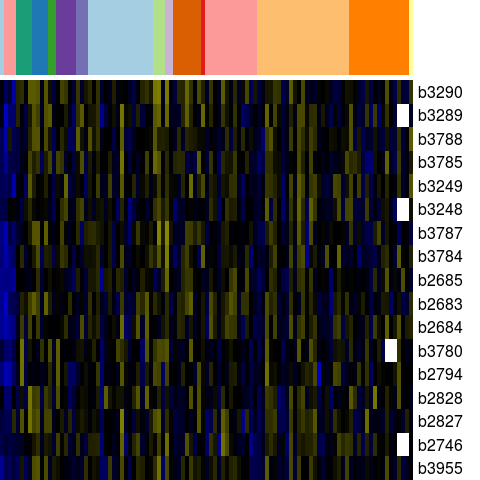
Genes in eco_16209
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b2683 | ygaH | NP_417168.1 | predicted inner membrane protein | 2808366 | 2808701 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2684 | mprA | NP_417169.1 | DNA-binding transcriptional repressor of microcin B17 synthesis and multidrug efflux | 2808792 | 2809322 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2685 | emrA | NP_417170.1 | multidrug efflux system | 2809449 | 2810621 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2746 | ispF | NP_417226.1 | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | 2869802 | 2869323 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2794 | yqcD | NP_417274.1 | hypothetical protein | 2923370 | 2924218 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2827 | thyA | NP_417304.1 | thymidylate synthase | 2963177 | 2962383 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2828 | lgt | NP_417305.1 | prolipoprotein diacylglyceryl transferase | 2964059 | 2963184 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3248 | yhdE | NP_417714.1 | Maf-like protein | 3396400 | 3395807 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3249 | mreD | NP_417715.1 | cell wall structural complex MreBCD transmembrane component MreD | 3396897 | 3396409 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3289 | rsmB | NP_417747.1 | 16S rRNA m5C967 methyltransferase, S-adenosyl-L-methionine-dependen | 3433229 | 3434518 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3290 | trkA | NP_417748.1 | potassium transporter peripheral membrane componen | 3434540 | 3435916 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3780 | rhlB | NP_418227.1 | ATP-dependent RNA helicase | 3963653 | 3962388 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3784 | rfe | NP_418231.1 | UDP-GlcNAc:undecaprenylphosphate GlcNAc-1-phosphate transferase | 3965939 | 3967042 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3785 | wzzE | NP_418232.2 | putative transport protein | 3967054 | 3968100 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3787 | rffD | YP_026254.1 | UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase | 3969283 | 3970545 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3788 | rffG | YP_026255.1 | dTDP-glucose 4,6-dehydratase | 3970545 | 3971612 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3955 | yijP | NP_418390.1 | conserved inner membrane protein | 4148288 | 4146555 | - | NC_000913.2 |
Conditions in eco_16209
| Condition ID | Condition Name |
|---|---|
| eco_109 | GSM1379 |
| eco_110 | GSM1380 |
| eco_111 | GSM18235 |
| eco_123 | GSM18279 |
| eco_14 | Clean_ANA7 |
| eco_142 | GSM63892 |
| eco_149 | GSM67647 |
| eco_160 | GSM73146 |
| eco_177 | GSM73277 |
| eco_193 | GSM82548 |
| eco_197 | GSM82616 |
| eco_223 | GSM99113 |
| eco_228 | GSM99128 |
| eco_232 | GSM99136 |
| eco_233 | GSM99150 |
| eco_237 | GSM99164 |
| eco_238 | GSM99167 |
| eco_245 | GSM99196 |
| eco_266 | 5265 |
| eco_272 | Clean_DIAUX9 |
| eco_279 | EC_GLY_GLY2_20_C |
| eco_284 | EC_LAC_LA_20_B |
| eco_286 | EC_LAC_LB_20_A |
| eco_289 | EC_LAC_LE_20_B |
| eco_294 | E-MEXP-245-raw-data-396455413.txt |
| eco_295 | E-MEXP-267-raw-data-495080191.txt |
| eco_306 | GSM101242 |
| eco_311 | GSM106340 |
| eco_323 | GSM118614 |
| eco_329 | GSM1374 |
| eco_330 | GSM1378 |
| eco_353 | GSM73137 |
| eco_358 | GSM73266 |
| eco_359 | GSM73269 |
| eco_368 | GSM8118 |
| eco_372 | GSM82553 |
| eco_373 | GSM82598 |
| eco_381 | GSM83081 |
| eco_392 | GSM99114 |
| eco_393 | GSM99118 |
| eco_395 | GSM99138 |
| eco_401 | GSM99162 |
| eco_402 | GSM99166 |
| eco_410 | GSM99204 |
| eco_42 | EC_GLY_GLYD_20_C |
| eco_422 | 5273 |
| eco_423 | 9395 |
| eco_429 | crpcysB |
| eco_458 | GSM1382 |
| eco_46 | EC_LAC_L2_60_C |
| eco_463 | GSM30819 |
| eco_464 | GSM35418 |
| eco_483 | GSM73136 |
| eco_486 | GSM73261 |
| eco_489 | GSM73282 |
| eco_491 | GSM77344 |
| eco_492 | GSM77348 |
| eco_500 | GSM88913 |
| eco_501 | GSM88915 |
| eco_502 | GSM98720 |
| eco_504 | GSM98728 |
| eco_51 | EC_LAC_LB_60_C |
| eco_517 | GSM99176 |
| eco_518 | GSM99188 |
| eco_52 | EC_LAC_LC_20_A |
| eco_529 | 1292 |
| eco_543 | EC_GLY_GLY1_20_B |
| eco_544 | EC_GLY_GLY2_44_A |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_558 | E-MEXP-267-raw-data-495080223.txt |
| eco_576 | GSM18261 |
| eco_583 | GSM63894 |
| eco_601 | GSM98730 |
| eco_607 | GSM99141 |
| eco_614 | GSM99210 |
| eco_617 | 1641 |
| eco_635 | EC_LAC_L3_20_B |
| eco_636 | EC_LAC_L3_60_B |
| eco_642 | E-MEXP-267-raw-data-495080335.txt |
| eco_651 | GSM18236 |
| eco_653 | GSM18274 |
| eco_663 | GSM65598 |
| eco_668 | GSM67658 |
| eco_670 | GSM73126 |
| eco_672 | GSM73148 |
| eco_695 | GSM99115 |
| eco_736 | GSM77346 |
| eco_738 | GSM82544 |
| eco_77 | GSM101240 |
| eco_776 | GSM77342 |
| eco_780 | GSM99147 |
| eco_789 | GSM101239 |
| eco_798 | GSM73253 |
| eco_811 | Clean_DIAUX17 |
| eco_815 | GSM101770 |
| eco_823 | EC_GLY_GLYB_44_B |
| eco_840 | GSM18272 |
| eco_851 | GSM99112 |
| eco_858 | GSM118609 |
| eco_860 | GSM82549 |
| eco_867 | GSM83075 |
| eco_868 | GSM73286 |
| eco_98 | GSM118605 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_16209_1 | 2700.0000 |  |
GREs assigned to eco_16209
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
Corems associated with eco_16209
| Corem |
|---|
| ec565697 |
| ec583534 |
| ec583538 |
| ec596323 |
| ec601661 |
| ec603570 |