eco_16242 Co-expression
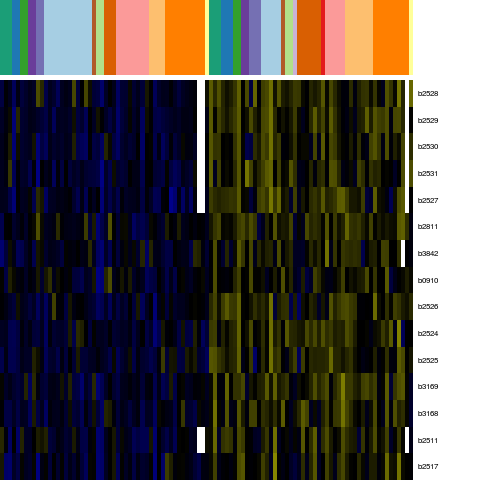
Genes in eco_16242
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0910 | cmk | NP_415430.1 | cytidylate kinase | 960424 | 961107 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2511 | b2511 | NP_417006.2 | putative GTP-binding factor | 2635378 | 2633906 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2517 | yfgB | NP_417012.1 | predicted enzyme | 2642305 | 2641151 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2524 | yfhJ | NP_417019.1 | hypothetical protein | 2654758 | 2654558 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2525 | fdx | NP_417020.1 | [2Fe-2S] ferredoxin | 2655105 | 2654770 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2526 | hscA | NP_417021.1 | chaperone protein HscA | 2656957 | 2655107 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2527 | hscB | NP_417022.1 | co-chaperone Hsc | 2657489 | 2656974 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2528 | iscA | NP_417023.1 | iron-sulfur cluster assembly protein | 2657908 | 2657585 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2529 | iscU | NP_417024.1 | scaffold protein | 2658311 | 2657925 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2530 | iscS | YP_026169.1 | cysteine desulfurase | 2659553 | 2658339 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2531 | iscR | NP_417026.1 | DNA-binding transcriptional repressor | 2660153 | 2659665 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2811 | ygdK | NP_417291.1 | predicted Fe-S metabolism protein | 2942564 | 2943007 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3168 | infB | NP_417637.1 | translation initiation factor IF-2 | 3314036 | 3311364 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3169 | nusA | NP_417638.1 | transcription elongation factor NusA | 3315548 | 3314061 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3842 | rfaH | NP_418284.1 | transcriptional activator RfaH | 4022844 | 4022356 | - | NC_000913.2 |
Conditions in eco_16242
| Condition ID | Condition Name |
|---|---|
| eco_109 | GSM1379 |
| eco_110 | GSM1380 |
| eco_123 | GSM18279 |
| eco_126 | GSM30817 |
| eco_14 | Clean_ANA7 |
| eco_194 | GSM82551 |
| eco_197 | GSM82616 |
| eco_212 | GSM98723 |
| eco_215 | GSM98732 |
| eco_223 | GSM99113 |
| eco_232 | GSM99136 |
| eco_233 | GSM99150 |
| eco_238 | GSM99167 |
| eco_249 | GSM99211 |
| eco_266 | 5265 |
| eco_27 | Clean_TGC18 |
| eco_278 | EC_GLU_WILD_C |
| eco_28 | Clean_TGC2 |
| eco_295 | E-MEXP-267-raw-data-495080191.txt |
| eco_298 | fur |
| eco_301 | GADXW_exp9 |
| eco_311 | GSM106340 |
| eco_314 | GSM106759 |
| eco_323 | GSM118614 |
| eco_325 | GSM118621 |
| eco_329 | GSM1374 |
| eco_330 | GSM1378 |
| eco_347 | GSM65624 |
| eco_358 | GSM73266 |
| eco_363 | GSM77358 |
| eco_372 | GSM82553 |
| eco_373 | GSM82598 |
| eco_392 | GSM99114 |
| eco_401 | GSM99162 |
| eco_402 | GSM99166 |
| eco_422 | 5273 |
| eco_423 | 9395 |
| eco_429 | crpcysB |
| eco_453 | GSM114385 |
| eco_458 | GSM1382 |
| eco_46 | EC_LAC_L2_60_C |
| eco_486 | GSM73261 |
| eco_491 | GSM77344 |
| eco_496 | GSM82600 |
| eco_5 | 1913 |
| eco_501 | GSM88915 |
| eco_502 | GSM98720 |
| eco_52 | EC_LAC_LC_20_A |
| eco_528 | wt__G |
| eco_529 | 1292 |
| eco_540 | crpfruR__G |
| eco_554 | EC_LAC_LD_20_A |
| eco_564 | GSM101780 |
| eco_573 | GSM137282 |
| eco_575 | GSM18253 |
| eco_576 | GSM18261 |
| eco_582 | GSM63891 |
| eco_589 | GSM77359 |
| eco_591 | GSM82608 |
| eco_601 | GSM98730 |
| eco_603 | GSM99087 |
| eco_614 | GSM99210 |
| eco_616 | 1278 |
| eco_633 | EC_GLY_WILD_E |
| eco_636 | EC_LAC_L3_60_B |
| eco_642 | E-MEXP-267-raw-data-495080335.txt |
| eco_653 | GSM18274 |
| eco_663 | GSM65598 |
| eco_668 | GSM67658 |
| eco_67 | E-MEXP-267-raw-data-495080207.txt |
| eco_672 | GSM73148 |
| eco_683 | GSM83063 |
| eco_687 | GSM88917 |
| eco_690 | GSM99082 |
| eco_705 | GSM99397 |
| eco_714 | EC_LAC_WILD_B |
| eco_736 | GSM77346 |
| eco_738 | GSM82544 |
| eco_740 | GSM89485 |
| eco_752 | GSM99182 |
| eco_780 | GSM99147 |
| eco_784 | crp__G |
| eco_789 | GSM101239 |
| eco_798 | GSM73253 |
| eco_80 | GSM101772 |
| eco_804 | GSM99093 |
| eco_805 | GSM99153 |
| eco_81 | GSM101773 |
| eco_811 | Clean_DIAUX17 |
| eco_815 | GSM101770 |
| eco_819 | GSM77352 |
| eco_823 | EC_GLY_GLYB_44_B |
| eco_826 | GSM106756 |
| eco_838 | GSM114615 |
| eco_840 | GSM18272 |
| eco_848 | GSM99639 |
| eco_851 | GSM99112 |
| eco_858 | GSM118609 |
| eco_86 | GSM111413 |
| eco_866 | GSM65601 |
| eco_867 | GSM83075 |
| eco_98 | GSM118605 |
| eco_99 | GSM118607 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_16242_1 | 410.0000 | 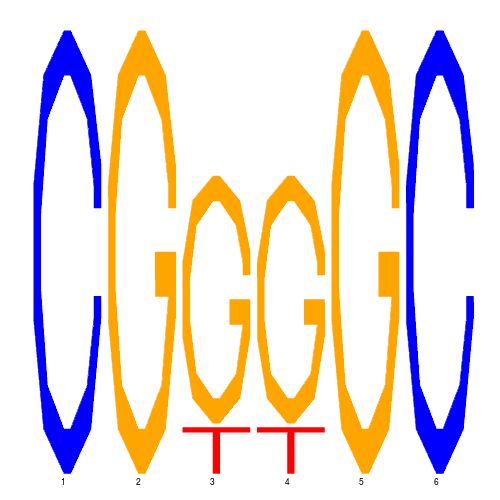 |
GREs assigned to eco_16242
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
Corems associated with eco_16242
| Corem |
|---|
| ec560740 |
| ec560742 |
| ec560788 |
| ec560793 |
| ec581127 |
| ec581148 |