eco_1776 Co-expression
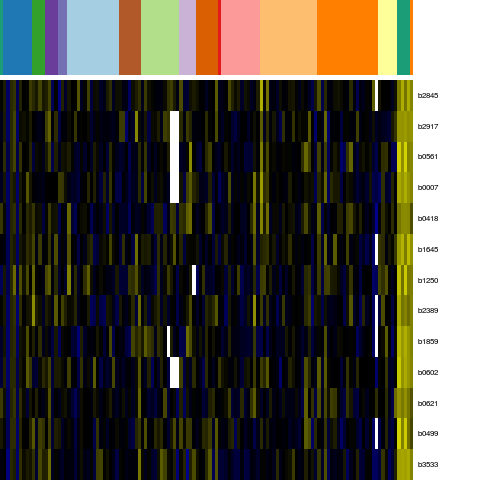
Genes in eco_1776
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0007 | yaaJ | NP_414548.1 | predicted transporter | 7959 | 6529 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0418 | pgpA | NP_414952.1 | phosphatidylglycerophosphatase A | 435813 | 436331 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0499 | ylbH | conserved protein, rhs-like | 527176 | 527883 | + | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b0561 | tfaD | orf, hypothetical protein | 580883 | 581320 | + | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b0602 | ybdN | NP_415135.1 | hypothetical protein | 635792 | 634572 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0621 | dcuC | NP_415154.1 | anaerobic C4-dicarboxylate transpor | 655191 | 653806 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1250 | kch | NP_415766.1 | voltage-gated potassium channe | 1308293 | 1307040 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1645 | ydhK | NP_416162.1 | conserved inner membrane protein | 1720145 | 1722157 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1859 | znuB | NP_416373.1 | high-affinity zinc transporter membrane componen | 1941438 | 1942223 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2389 | yfeO | NP_416890.1 | hypothetical protein | 2507652 | 2508908 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2845 | yqeG | NP_417322.1 | predicted transporter | 2983869 | 2985098 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2917 | yliK | NP_417392.1 | methylmalonyl-CoA mutase | 3058872 | 3061016 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3533 | yhjO | NP_417990.4 | putative cellulose synthase | 3693259 | 3690641 | - | NC_000913.2 |
Conditions in eco_1776
| Condition ID | Condition Name |
|---|---|
| eco_10 | 8455 |
| eco_104 | GSM120192 |
| eco_116 | GSM18256 |
| eco_117 | GSM18257 |
| eco_119 | GSM18262 |
| eco_149 | GSM67647 |
| eco_15 | Clean_DIAUX11 |
| eco_18 | Clean_DIAUX3 |
| eco_185 | GSM77354 |
| eco_188 | GSM8116 |
| eco_191 | GSM82542 |
| eco_200 | GSM83060 |
| eco_202 | GSM83065 |
| eco_21 | Clean_H2O211 |
| eco_210 | GSM98717 |
| eco_220 | GSM99107 |
| eco_246 | GSM99197 |
| eco_254 | GSM99395 |
| eco_258 | 1277 |
| eco_260 | 1592 |
| eco_264 | 1644 |
| eco_267 | 5278 |
| eco_268 | 5284 |
| eco_286 | EC_LAC_LB_20_A |
| eco_294 | E-MEXP-245-raw-data-396455413.txt |
| eco_3 | 1593 |
| eco_300 | GADXW_exp8 |
| eco_310 | GSM106339 |
| eco_325 | GSM118621 |
| eco_327 | GSM1367 |
| eco_358 | GSM73266 |
| eco_361 | GSM73281 |
| eco_366 | GSM8115 |
| eco_368 | GSM8118 |
| eco_369 | GSM82543 |
| eco_380 | GSM83074 |
| eco_383 | GSM88912 |
| eco_385 | GSM98718 |
| eco_386 | GSM98731 |
| eco_395 | GSM99138 |
| eco_396 | GSM99143 |
| eco_397 | GSM99145 |
| eco_403 | GSM99172 |
| eco_405 | GSM99187 |
| eco_415 | GSM99400 |
| eco_425 | Clean_TGC12 |
| eco_429 | crpcysB |
| eco_44 | EC_LAC_L2_20_C |
| eco_451 | GSM114371 |
| eco_453 | GSM114385 |
| eco_455 | GSM118617 |
| eco_458 | GSM1382 |
| eco_468 | GSM37885 |
| eco_471 | GSM62198 |
| eco_473 | GSM65588 |
| eco_475 | GSM65623 |
| eco_478 | GSM67651 |
| eco_483 | GSM73136 |
| eco_487 | GSM73268 |
| eco_488 | GSM73279 |
| eco_490 | GSM73290 |
| eco_507 | GSM99088 |
| eco_508 | GSM99092 |
| eco_511 | GSM99104 |
| eco_524 | GSM99394 |
| eco_529 | 1292 |
| eco_530 | 14830 |
| eco_538 | crp |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_552 | EC_LAC_LB_60_A |
| eco_559 | E-MEXP-267-raw-data-495080239.txt |
| eco_567 | GSM111423 |
| eco_59 | E-MEXP-222-raw-data-386284924.txt |
| eco_591 | GSM82608 |
| eco_608 | GSM99151 |
| eco_631 | EC_GLY_GLYE_44_A |
| eco_646 | GSM116027 |
| eco_663 | GSM65598 |
| eco_665 | GSM65622 |
| eco_678 | GSM77356 |
| eco_680 | GSM8114 |
| eco_682 | GSM83054 |
| eco_688 | GSM89487 |
| eco_689 | GSM98729 |
| eco_693 | GSM99100 |
| eco_695 | GSM99115 |
| eco_697 | GSM99144 |
| eco_698 | GSM99155 |
| eco_703 | GSM99216 |
| eco_709 | Clean_H2O29 |
| eco_719 | GSM120196 |
| eco_720 | GSM1369 |
| eco_722 | GSM30816 |
| eco_734 | GSM73252 |
| eco_736 | GSM77346 |
| eco_738 | GSM82544 |
| eco_741 | GSM89486 |
| eco_758 | EC_GLY_GLYD_44_C |
| eco_760 | EC_LAC_LD_60_B |
| eco_762 | GADXW_exp10 |
| eco_763 | GADXW_exp5 |
| eco_766 | GSM116031 |
| eco_77 | GSM101240 |
| eco_777 | GSM83059 |
| eco_783 | Clean_DIAUX8 |
| eco_785 | EC_LAC_L3_20_A |
| eco_793 | GSM116029 |
| eco_798 | GSM73253 |
| eco_804 | GSM99093 |
| eco_806 | GSM99175 |
| eco_810 | 5272 |
| eco_817 | GSM18277 |
| eco_822 | GSM99154 |
| eco_823 | EC_GLY_GLYB_44_B |
| eco_825 | GADXW_exp3 |
| eco_828 | GSM65596 |
| eco_829 | GSM67663 |
| eco_834 | GSM99212 |
| eco_838 | GSM114615 |
| eco_841 | GSM37076 |
| eco_845 | GSM118604 |
| eco_850 | GSM88918 |
| eco_851 | GSM99112 |
| eco_86 | GSM111413 |
| eco_864 | GSM99158 |
| eco_865 | Clean_DIAUX6 |
| eco_89 | GSM111420 |
| eco_9 | 5287 |
| eco_94 | GSM114614 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_1776_1 | 1100.0000 | 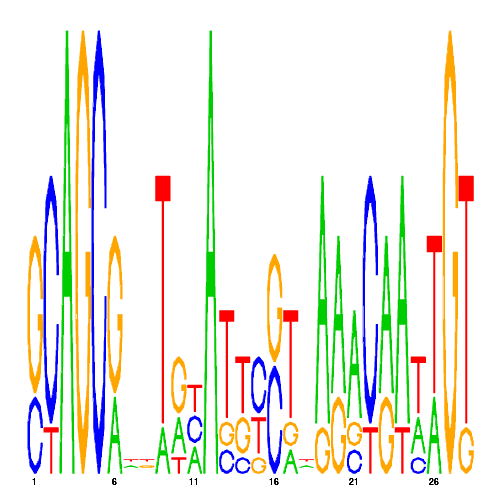 |
GREs assigned to eco_1776
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
Corems associated with eco_1776
| Corem |
|---|