eco_3426 Co-expression
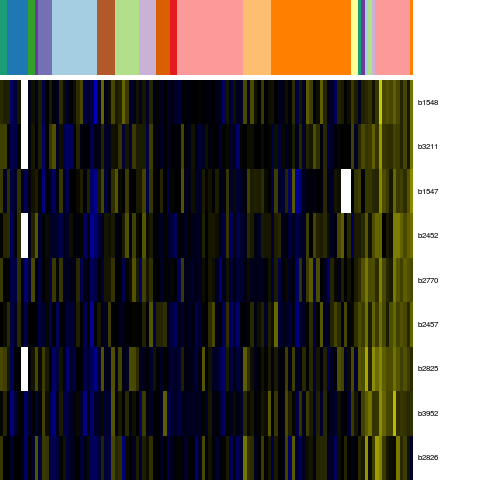
Genes in eco_3426
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1547 | stfQ | NP_416065.1 | Qin prophage; predicted side tail fibre assembly protein | 1633871 | 1632909 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1548 | nohA | Qin prophage; predicted packaging protein | 1634391 | 1633864 | - | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b2452 | eutH | NP_416947.1 | predicted inner membrane protein | 2566129 | 2564903 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2457 | cchA | NP_416952.2 | detox protein | 2570472 | 2570179 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2770 | ygcR | NP_417250.4 | putative transport protein | 2894577 | 2893798 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2825 | ppdB | NP_417302.1 | hypothetical protein | 2961738 | 2961175 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2826 | ppdA | NP_417303.1 | hypothetical protein | 2962199 | 2961729 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3211 | yhcC | NP_417678.1 | predicted Fe-S oxidoreductase | 3352072 | 3351143 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3952 | pflC | NP_418387.3 | probable pyruvate formate lyase activating enzyme 2 | 4144281 | 4145159 | + | NC_000913.2 |
Conditions in eco_3426
| Condition ID | Condition Name |
|---|---|
| eco_102 | GSM118620 |
| eco_127 | GSM30820 |
| eco_129 | GSM35417 |
| eco_137 | GSM62194 |
| eco_141 | GSM63889 |
| eco_142 | GSM63892 |
| eco_149 | GSM67647 |
| eco_150 | GSM67656 |
| eco_196 | GSM82613 |
| eco_199 | GSM83058 |
| eco_205 | GSM83077 |
| eco_208 | GSM88914 |
| eco_210 | GSM98717 |
| eco_223 | GSM99113 |
| eco_23 | Clean_H2O24 |
| eco_234 | GSM99152 |
| eco_246 | GSM99197 |
| eco_261 | 1595 |
| eco_262 | 1638 |
| eco_267 | 5278 |
| eco_273 | Clean_H2O23 |
| eco_276 | cysB__G |
| eco_280 | EC_GLY_GLYA_20_B |
| eco_282 | EC_GLY_GLYE_20_A |
| eco_288 | EC_LAC_LD_20_B |
| eco_295 | E-MEXP-267-raw-data-495080191.txt |
| eco_30 | cysB |
| eco_304 | GSM101237 |
| eco_306 | GSM101242 |
| eco_31 | EC_GLY_GLY1_20_C |
| eco_314 | GSM106759 |
| eco_316 | GSM111419 |
| eco_317 | GSM114370 |
| eco_321 | GSM118611 |
| eco_330 | GSM1378 |
| eco_34 | EC_GLY_GLY2_20_A |
| eco_346 | GSM65593 |
| eco_347 | GSM65624 |
| eco_356 | GSM73260 |
| eco_358 | GSM73266 |
| eco_36 | EC_GLY_GLYA_20_C |
| eco_366 | GSM8115 |
| eco_367 | GSM8117 |
| eco_37 | EC_GLY_GLYA_44_B |
| eco_394 | GSM99125 |
| eco_396 | GSM99143 |
| eco_405 | GSM99187 |
| eco_410 | GSM99204 |
| eco_429 | crpcysB |
| eco_432 | EC_GLY_GLYC_44_C |
| eco_433 | EC_LAC_L3_20_C |
| eco_44 | EC_LAC_L2_20_C |
| eco_447 | GSM106342 |
| eco_465 | GSM37068 |
| eco_467 | GSM37074 |
| eco_469 | GSM38546 |
| eco_470 | GSM38548 |
| eco_471 | GSM62198 |
| eco_477 | GSM65634 |
| eco_480 | GSM67659 |
| eco_494 | GSM77355 |
| eco_496 | GSM82600 |
| eco_498 | GSM82610 |
| eco_5 | 1913 |
| eco_500 | GSM88913 |
| eco_503 | GSM98725 |
| eco_529 | 1292 |
| eco_53 | EC_LAC_LC_60_A |
| eco_543 | EC_GLY_GLY1_20_B |
| eco_558 | E-MEXP-267-raw-data-495080223.txt |
| eco_575 | GSM18253 |
| eco_579 | GSM37063 |
| eco_591 | GSM82608 |
| eco_598 | GSM94002 |
| eco_599 | GSM94003 |
| eco_600 | GSM98719 |
| eco_610 | GSM99174 |
| eco_615 | GSM99399 |
| eco_62 | E-MEXP-222-raw-data-386285101.txt |
| eco_624 | Clean_TGC1 |
| eco_626 | EC_GLY_GLY2_20_B |
| eco_627 | EC_GLY_GLY2_44_B |
| eco_631 | EC_GLY_GLYE_44_A |
| eco_634 | EC_LAC_L2_20_A |
| eco_64 | E-MEXP-245-raw-data-396455333.txt |
| eco_641 | E-MEXP-244-raw-data-396455050.txt |
| eco_642 | E-MEXP-267-raw-data-495080335.txt |
| eco_646 | GSM116027 |
| eco_649 | GSM118610 |
| eco_650 | GSM1376 |
| eco_657 | GSM30818 |
| eco_661 | GSM63885 |
| eco_667 | GSM67657 |
| eco_669 | GSM67665 |
| eco_685 | GSM83068 |
| eco_711 | EC_GLY_GLYE_20_C |
| eco_718 | GSM118608 |
| eco_728 | GSM65597 |
| eco_742 | GSM94004 |
| eco_750 | GSM99178 |
| eco_753 | 5281 |
| eco_757 | EC_GLY_GLYC_44_B |
| eco_759 | EC_GLY_WILD_C |
| eco_760 | EC_LAC_LD_60_B |
| eco_765 | GSM114612 |
| eco_766 | GSM116031 |
| eco_768 | GSM18278 |
| eco_777 | GSM83059 |
| eco_782 | GSM99209 |
| eco_788 | GSM101238 |
| eco_82 | GSM106337 |
| eco_823 | EC_GLY_GLYB_44_B |
| eco_829 | GSM67663 |
| eco_836 | EC_GLY_GLYD_20_B |
| eco_838 | GSM114615 |
| eco_839 | GSM1370 |
| eco_847 | GSM73135 |
| eco_85 | GSM106758 |
| eco_860 | GSM82549 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_3426_1 | 7.6000 | 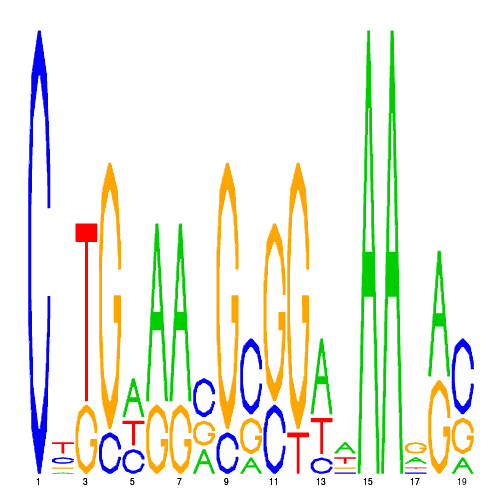 |
| eco_3426_2 | 1100.0000 | 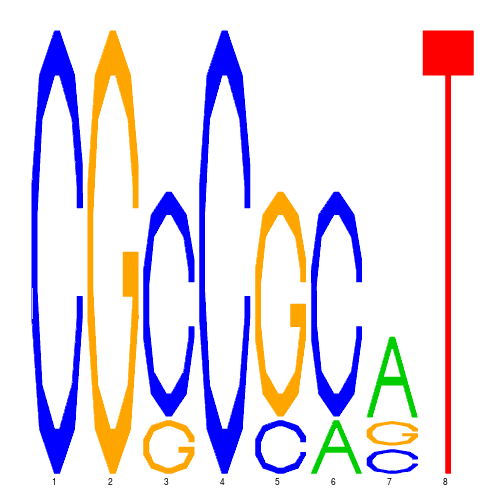 |
GREs assigned to eco_3426
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_0 |  |
Corems associated with eco_3426
| Corem |
|---|