eco_40626 Co-expression
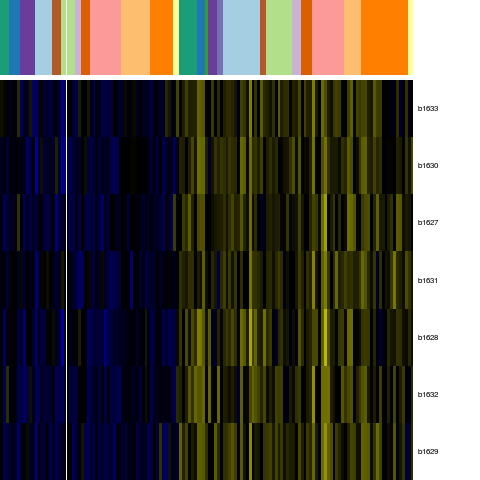
Genes in eco_40626
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1627 | rsxA | NP_416144.1 | Na(+)-translocating NADH-quinone reductase subunit E | 1703791 | 1704372 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1628 | rsxB | NP_416145.1 | electron transport complex protein Rn | 1704372 | 1704950 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1629 | rsxC | NP_416146.1 | electron transport complex protein Rn | 1704943 | 1707165 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1630 | rsxD | NP_416147.1 | electron transport complex protein RnfD | 1707166 | 1708224 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1631 | rsxG | NP_416148.1 | electron transport complex protein RnfG | 1708228 | 1708848 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1632 | rsxE | NP_416149.1 | NADH-ubiquinone oxidoreductase | 1708852 | 1709547 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1633 | nth | NP_416150.1 | DNA glycosylase and apyrimidinic (AP) lyase (endonuclease III) | 1709547 | 1710182 | + | NC_000913.2 |
Conditions in eco_40626
| Condition ID | Condition Name |
|---|---|
| eco_10 | 8455 |
| eco_100 | GSM118612 |
| eco_103 | GSM120166 |
| eco_104 | GSM120192 |
| eco_106 | GSM1371 |
| eco_112 | GSM18247 |
| eco_118 | GSM18260 |
| eco_138 | GSM62195 |
| eco_140 | GSM63887 |
| eco_141 | GSM63889 |
| eco_142 | GSM63892 |
| eco_144 | GSM65590 |
| eco_167 | GSM73160 |
| eco_195 | GSM82603 |
| eco_200 | GSM83060 |
| eco_211 | GSM98721 |
| eco_216 | GSM99090 |
| eco_221 | GSM99108 |
| eco_231 | GSM99133 |
| eco_232 | GSM99136 |
| eco_233 | GSM99150 |
| eco_249 | GSM99211 |
| eco_253 | GSM99393 |
| eco_254 | GSM99395 |
| eco_261 | 1595 |
| eco_276 | cysB__G |
| eco_295 | E-MEXP-267-raw-data-495080191.txt |
| eco_297 | E-MEXP-267-raw-data-495080367.txt |
| eco_298 | fur |
| eco_302 | GSM101231 |
| eco_305 | GSM101241 |
| eco_306 | GSM101242 |
| eco_310 | GSM106339 |
| eco_317 | GSM114370 |
| eco_320 | GSM116030 |
| eco_328 | GSM1372 |
| eco_329 | GSM1374 |
| eco_330 | GSM1378 |
| eco_331 | GSM1381 |
| eco_336 | GSM18275 |
| eco_34 | EC_GLY_GLY2_20_A |
| eco_348 | GSM65636 |
| eco_355 | GSM73255 |
| eco_357 | GSM73262 |
| eco_364 | GSM77360 |
| eco_367 | GSM8117 |
| eco_374 | GSM82604 |
| eco_380 | GSM83074 |
| eco_381 | GSM83081 |
| eco_385 | GSM98718 |
| eco_39 | EC_GLY_GLYB_44_A |
| eco_391 | GSM99105 |
| eco_399 | GSM99149 |
| eco_402 | GSM99166 |
| eco_416 | 1290 |
| eco_419 | 1637 |
| eco_421 | 5266 |
| eco_423 | 9395 |
| eco_424 | Clean_ANA2 |
| eco_429 | crpcysB |
| eco_43 | EC_GLY_WILD_D |
| eco_431 | EC_GLY_GLY2_44_C |
| eco_435 | EC_LAC_LC_20_C |
| eco_446 | GSM101769 |
| eco_449 | GSM111414 |
| eco_460 | GSM18259 |
| eco_483 | GSM73136 |
| eco_489 | GSM73282 |
| eco_49 | EC_LAC_LA_60_D |
| eco_500 | GSM88913 |
| eco_504 | GSM98728 |
| eco_511 | GSM99104 |
| eco_513 | GSM99134 |
| eco_514 | GSM99135 |
| eco_524 | GSM99394 |
| eco_533 | 1916 |
| eco_538 | crp |
| eco_540 | crpfruR__G |
| eco_542 | EC_GLU_WILD_B |
| eco_551 | EC_LAC_LA_60_C |
| eco_560 | E-MEXP-267-raw-data-495080303.txt |
| eco_563 | GSM101778 |
| eco_567 | GSM111423 |
| eco_570 | GSM118618 |
| eco_575 | GSM18253 |
| eco_579 | GSM37063 |
| eco_59 | E-MEXP-222-raw-data-386284924.txt |
| eco_592 | GSM83056 |
| eco_600 | GSM98719 |
| eco_603 | GSM99087 |
| eco_607 | GSM99141 |
| eco_608 | GSM99151 |
| eco_616 | 1278 |
| eco_617 | 1641 |
| eco_620 | Clean_DIAUX14 |
| eco_621 | Clean_DIAUX15 |
| eco_623 | Clean_H2O21 |
| eco_626 | EC_GLY_GLY2_20_B |
| eco_638 | EC_LAC_LB_60_B |
| eco_641 | E-MEXP-244-raw-data-396455050.txt |
| eco_645 | GSM106338 |
| eco_659 | GSM37066 |
| eco_662 | GSM63890 |
| eco_664 | GSM65600 |
| eco_669 | GSM67665 |
| eco_680 | GSM8114 |
| eco_681 | GSM82546 |
| eco_682 | GSM83054 |
| eco_683 | GSM83063 |
| eco_690 | GSM99082 |
| eco_698 | GSM99155 |
| eco_700 | GSM99179 |
| eco_703 | GSM99216 |
| eco_707 | Clean_ANA15 |
| eco_708 | Clean_ANA9 |
| eco_730 | GSM67655 |
| eco_736 | GSM77346 |
| eco_746 | GSM99110 |
| eco_75 | GSM101234 |
| eco_76 | GSM101236 |
| eco_760 | EC_LAC_LD_60_B |
| eco_761 | EC_LAC_WILD_A |
| eco_772 | GSM73143 |
| eco_775 | GSM73289 |
| eco_776 | GSM77342 |
| eco_78 | GSM101244 |
| eco_783 | Clean_DIAUX8 |
| eco_785 | EC_LAC_L3_20_A |
| eco_787 | fruR__G |
| eco_788 | GSM101238 |
| eco_791 | GSM114383 |
| eco_792 | GSM114613 |
| eco_795 | GSM18246 |
| eco_817 | GSM18277 |
| eco_818 | GSM73129 |
| eco_82 | GSM106337 |
| eco_822 | GSM99154 |
| eco_826 | GSM106756 |
| eco_841 | GSM37076 |
| eco_849 | GSM120197 |
| eco_852 | GSM99156 |
| eco_86 | GSM111413 |
| eco_87 | GSM111415 |
| eco_88 | GSM111417 |
| eco_90 | GSM111421 |
| eco_92 | GSM114368 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_40626_2 | 9999999.0000 |  |
| eco_40626_1 | 9999999.0000 |  |
GREs assigned to eco_40626
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_0 |  |
Corems associated with eco_40626
| Corem |
|---|
| ec537196 |
| ec537197 |
| ec537998 |
| ec560748 |
| ec583533 |