eco_43645 Co-expression
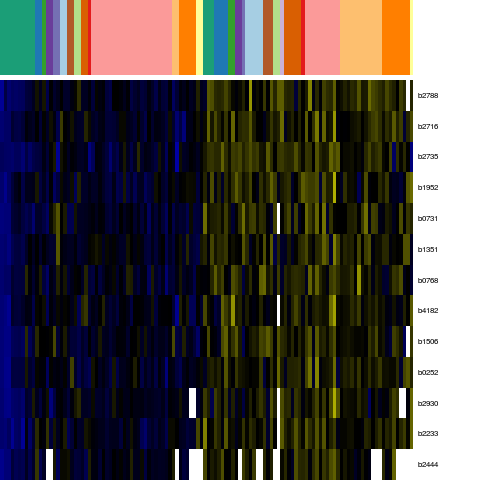
Genes in eco_43645
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0252 | yafZ | NP_414786.4 | orf, hypothetical protein | 267229 | 266408 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0731 | mngA | NP_415259.1 | fused 2-O-a-mannosyl-D-glycerate specific PTS enzymes: IIA component/IIB component/IIC componen | 765207 | 767183 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0768 | ybhD | NP_415289.4 | predicted DNA-binding transcriptional regulator | 799798 | 798845 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1351 | racC | NP_415867.1 | Rac prophage; predicted protein | 1415787 | 1415512 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1506 | yneL | predicted transcriptional regulator | 1588560 | 1588358 | - | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b1952 | dsrB | NP_416462.1 | hypothetical protein | 2022847 | 2022659 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2233 | yfaL | NP_416736.1 | adhesin | 2342191 | 2338439 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2444 | yffM | NP_416939.1 | CPZ-55 prophage; predicted protein | 2559390 | 2559635 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2716 | ascB | NP_417196.1 | cryptic 6-phospho-beta-glucosidase | 2839012 | 2840436 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2735 | ygbI | NP_417215.2 | predicted DNA-binding transcriptional regulator | 2859256 | 2858489 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2788 | gudX | NP_417268.1 | predicted glucarate dehydratase | 2918768 | 2917428 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2930 | yggF | NP_417405.1 | predicted hexoseP phosphatase | 3074204 | 3073239 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b4182 | yjfJ | NP_418603.1 | predicted transcriptional regulator effector protein | 4408576 | 4409274 | + | NC_000913.2 |
Conditions in eco_43645
| Condition ID | Condition Name |
|---|---|
| eco_125 | GSM30815 |
| eco_127 | GSM30820 |
| eco_136 | GSM38547 |
| eco_138 | GSM62195 |
| eco_140 | GSM63887 |
| eco_163 | GSM73155 |
| eco_173 | GSM73270 |
| eco_19 | Clean_DIAUX7 |
| eco_2 | 1285 |
| eco_211 | GSM98721 |
| eco_216 | GSM99090 |
| eco_217 | GSM99091 |
| eco_218 | GSM99096 |
| eco_219 | GSM99103 |
| eco_222 | GSM99109 |
| eco_225 | GSM99119 |
| eco_230 | GSM99132 |
| eco_242 | GSM99186 |
| eco_246 | GSM99197 |
| eco_255 | GSM99396 |
| eco_264 | 1644 |
| eco_274 | Clean_H2O26 |
| eco_282 | EC_GLY_GLYE_20_A |
| eco_287 | EC_LAC_LB_20_C |
| eco_288 | EC_LAC_LD_20_B |
| eco_290 | EC_LAC_WILD_D |
| eco_303 | GSM101235 |
| eco_307 | GSM101775 |
| eco_309 | GSM101779 |
| eco_313 | GSM106753 |
| eco_319 | GSM116025 |
| eco_32 | EC_GLY_GLY1_44_A |
| eco_329 | GSM1374 |
| eco_331 | GSM1381 |
| eco_340 | GSM23467 |
| eco_376 | GSM83053 |
| eco_379 | GSM83073 |
| eco_399 | GSM99149 |
| eco_400 | GSM99157 |
| eco_41 | EC_GLY_GLYC_20_C |
| eco_417 | 14829 |
| eco_42 | EC_GLY_GLYD_20_C |
| eco_421 | 5266 |
| eco_427 | Clean_TGC6 |
| eco_433 | EC_LAC_L3_20_C |
| eco_439 | GADXW_exp1 |
| eco_44 | EC_LAC_L2_20_C |
| eco_440 | GADXW_exp11 |
| eco_441 | GADXW_exp14 |
| eco_443 | GADXW_exp16 |
| eco_445 | GSM101768 |
| eco_45 | EC_LAC_L2_60_B |
| eco_451 | GSM114371 |
| eco_464 | GSM35418 |
| eco_470 | GSM38548 |
| eco_477 | GSM65634 |
| eco_48 | EC_LAC_LA_60_A |
| eco_486 | GSM73261 |
| eco_492 | GSM77348 |
| eco_500 | GSM88913 |
| eco_524 | GSM99394 |
| eco_530 | 14830 |
| eco_538 | crp |
| eco_539 | crpcysB__G |
| eco_544 | EC_GLY_GLY2_44_A |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_55 | EC_LAC_LE_60_A |
| eco_554 | EC_LAC_LD_20_A |
| eco_561 | E-MEXP-267-raw-data-495080319.txt |
| eco_564 | GSM101780 |
| eco_569 | GSM118616 |
| eco_580 | GSM37887 |
| eco_587 | GSM77345 |
| eco_588 | GSM77349 |
| eco_589 | GSM77359 |
| eco_590 | GSM77361 |
| eco_601 | GSM98730 |
| eco_603 | GSM99087 |
| eco_628 | EC_GLY_GLYB_20_A |
| eco_630 | EC_GLY_GLYC_44_A |
| eco_631 | EC_GLY_GLYE_44_A |
| eco_632 | EC_GLY_WILD_B |
| eco_635 | EC_LAC_L3_20_B |
| eco_636 | EC_LAC_L3_60_B |
| eco_639 | EC_LAC_LC_60_B |
| eco_640 | EC_LAC_LE_60_B |
| eco_688 | GSM89487 |
| eco_701 | GSM99190 |
| eco_704 | GSM99380 |
| eco_705 | GSM99397 |
| eco_709 | Clean_H2O29 |
| eco_71 | GADXW_exp6 |
| eco_712 | EC_LAC_LA_20_A |
| eco_718 | GSM118608 |
| eco_73 | GSM101232 |
| eco_731 | GSM67660 |
| eco_747 | GSM99120 |
| eco_750 | GSM99178 |
| eco_752 | GSM99182 |
| eco_753 | 5281 |
| eco_754 | Clean_ANA17 |
| eco_755 | Clean_DIAUX2 |
| eco_757 | EC_GLY_GLYC_44_B |
| eco_758 | EC_GLY_GLYD_44_C |
| eco_760 | EC_LAC_LD_60_B |
| eco_761 | EC_LAC_WILD_A |
| eco_762 | GADXW_exp10 |
| eco_763 | GADXW_exp5 |
| eco_766 | GSM116031 |
| eco_770 | GSM37073 |
| eco_772 | GSM73143 |
| eco_773 | GSM73153 |
| eco_775 | GSM73289 |
| eco_777 | GSM83059 |
| eco_778 | GSM88919 |
| eco_94 | GSM114614 |
| eco_96 | GSM116026 |
| eco_99 | GSM118607 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_43645_1 | 34.0000 | 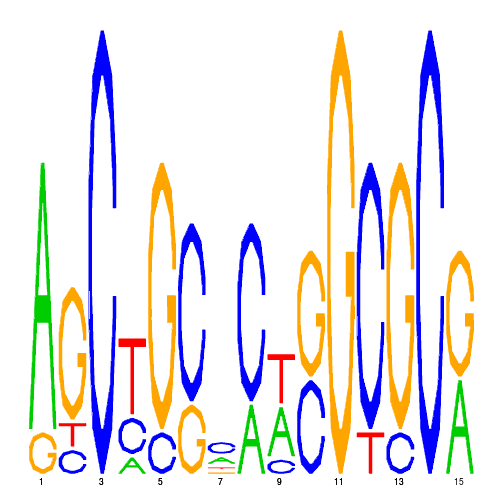 |
| eco_43645_2 | 260.0000 | 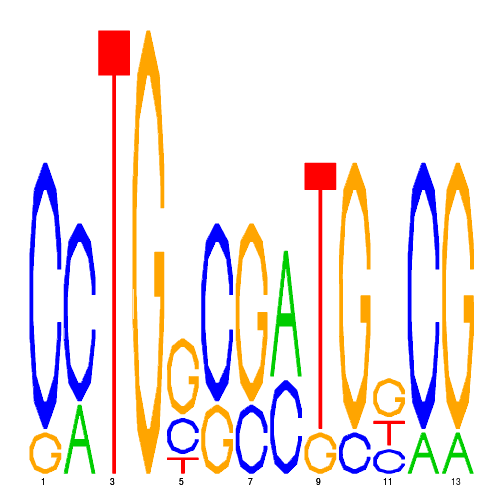 |
GREs assigned to eco_43645
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_0 |  |
Corems associated with eco_43645
| Corem |
|---|