hal_1208 Co-expression
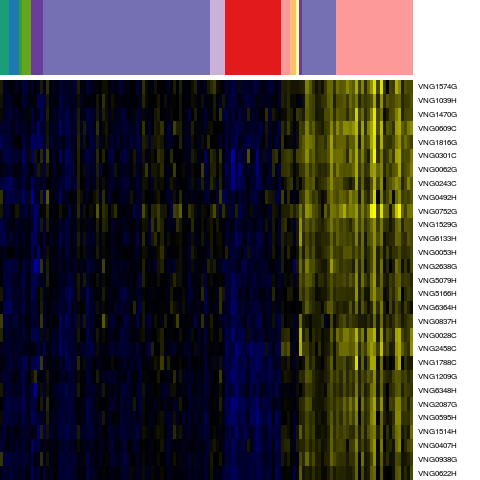
Genes in hal_1208
Conditions in hal_1208
| Condition ID | Condition Name |
|---|---|
| hal_1 | light2__1440m_vs_light1_3960m |
| hal_10 | light2__3060m_vs_light1_3960m |
| hal_1060 | o2_set1_0001m_L2H_500_vs_NRC-1c |
| hal_1061 | o2_set1_0005m_H_922_vs_NRC-1c |
| hal_1062 | o2_set1_0010m_H_947_vs_NRC-1c |
| hal_1063 | o2_set1_0020m_H_937_vs_NRC-1c |
| hal_1064 | o2_set1_0030m_H_931_vs_NRC-1c |
| hal_1065 | o2_set1_0045m_H_939_vs_NRC-1c |
| hal_1066 | o2_set1_0090m_H_941_vs_NRC-1c |
| hal_1071 | o2_set1_0372m_H2L_079_vs_NRC-1c |
| hal_1073 | o2_set1_0387m_L_041_vs_NRC-1c |
| hal_1074 | o2_set1_0392m_L_037_vs_NRC-1c |
| hal_1075 | o2_set1_0397m_L_035_vs_NRC-1c |
| hal_1076 | o2_set1_0407m_L_030_vs_NRC-1c |
| hal_1078 | o2_set1_0432m_L_024_vs_NRC-1c |
| hal_1080 | o2_set1_0567m_L_011_vs_NRC-1c |
| hal_1081 | o2_set1_0747m_L_008_vs_NRC-1c |
| hal_1082 | o2_set1_0750m_L2H_540_vs_NRC-1c |
| hal_1083 | o2_set1_0755m_L2H_825_vs_NRC-1c |
| hal_1084 | o2_set1_0760m_L2H_847_vs_NRC-1c |
| hal_1086 | o2_set1_0780m_L2H_847_vs_NRC-1c |
| hal_18 | light2__4500m_vs_light1_3960m |
| hal_24 | light2__5580m_vs_light1_3960m |
| hal_27 | dark2__0060m_vs_NRC-1c |
| hal_275 | zn__0005um_vs_NRC-1 |
| hal_276 | zn__0010um_vs_NRC-1 |
| hal_277 | zn__0020um_vs_NRC-1 |
| hal_279 | cu__0850um_vs_NRC-1 |
| hal_28 | dark2__0300m_vs_NRC-1c |
| hal_30 | dark2__0780m_vs_NRC-1c |
| hal_31 | dark2__1020m_vs_NRC-1c |
| hal_32 | dark2__1260m_vs_NRC-1c |
| hal_33 | dark2__1500m_vs_NRC-1c |
| hal_34 | dark2__1740m_vs_NRC-1c |
| hal_35 | dark2__1980m_vs_NRC-1c |
| hal_37 | dark2__2460m_vs_NRC-1c |
| hal_39 | dark2__2940m_vs_NRC-1c |
| hal_40 | dark2__3180m_vs_NRC-1c |
| hal_41 | dark2__3420m_vs_NRC-1c |
| hal_42 | dark2__3660m_vs_NRC-1c |
| hal_434 | ni__0000um_vs_NRC-1c |
| hal_435 | ni__0500um_vs_NRC-1c |
| hal_437 | ni__1500um_vs_NRC-1c |
| hal_438 | mn__0800um_vs_NRC-1 |
| hal_45 | dark1__0240m_vs_NRC-1c |
| hal_458 | fe__2000um_vs_NRC-1 |
| hal_459 | fe__4000um_vs_NRC-1 |
| hal_46 | dark1__0480m_vs_NRC-1c |
| hal_464 | feso4__0005m_vs_NRC-1b |
| hal_465 | feso4__0010m_vs_NRC-1b |
| hal_47 | dark1__0720m_vs_NRC-1c |
| hal_471 | feso4__0160m_vs_NRC-1 |
| hal_48 | dark1__0960m_vs_NRC-1c |
| hal_481 | co__0000um_vs_NRC-1c |
| hal_483 | co__0300um_vs_NRC-1c |
| hal_484 | co__0500um_vs_NRC-1c |
| hal_49 | dark1__1200m_vs_NRC-1c |
| hal_494 | VNG2334C__LO_L_vs_NRC-1 |
| hal_495 | VNG1296C__HO_D_vs_NRC-1 |
| hal_496 | VNG1296C__HO_L_vs_NRC-1 |
| hal_5 | light2__2160m_vs_light1_3960m |
| hal_50 | dark1__1440m_vs_NRC-1c |
| hal_502 | VNG0750C__LO_L_vs_NRC-1 |
| hal_506 | VNG0019H__LO_L_vs_NRC-1c |
| hal_507 | uv__000jm2_CL_060m |
| hal_508 | ura3__HO_D_vs_NRC-1 |
| hal_509 | ura3__HO_L_vs_NRC-1 |
| hal_511 | ura3__LO_L_vs_NRC-1 |
| hal_513 | tfb_D_vs_NRC-1c |
| hal_518 | tbp_C_vs_NRC-1c |
| hal_52 | dark1__1920m_vs_NRC-1c |
| hal_520 | sop2_II__HO_D_vs_NRC-1c |
| hal_522 | sop2_II__LO_D_vs_NRC-1c |
| hal_527 | sop2_I__LO_L_vs_NRC-1 |
| hal_531 | phr2__LO_L_vs_NRC-1 |
| hal_532 | phr1_and_2__HO_D_vs_NRC-1 |
| hal_533 | phr1_and_2__HO_L_vs_NRC-1 |
| hal_534 | phr1_and_2__LO_D_vs_NRC-1 |
| hal_535 | phr1_and_2__LO_L_vs_NRC-1 |
| hal_536 | phr1__HO_D_vs_NRC-1 |
| hal_539 | phr1__LO_L_vs_NRC-1 |
| hal_54 | dark1__2400m_vs_NRC-1c |
| hal_541 | phoR__HO_L_vs_NRC-1 |
| hal_542 | phoR__LO_D_vs_NRC-1 |
| hal_543 | phoR__LO_L_vs_NRC-1 |
| hal_55 | dark1__2640m_vs_NRC-1c |
| hal_555 | kinA2_II__HO_D_vs_NRC-1c |
| hal_556 | kinA2_II__HO_L_vs_NRC-1c |
| hal_557 | kinA2_II__LO_D_vs_NRC-1c |
| hal_56 | dark1__2880m_vs_NRC-1c |
| hal_562 | kinA2_I__LO_L_vs_NRC-1 |
| hal_563 | htr8__HO_D_vs_NRC-1 |
| hal_565 | htr8__LO_D_vs_NRC-1 |
| hal_569 | htr2__LO_D_vs_NRC-1 |
| hal_57 | dark1__3120m_vs_NRC-1c |
| hal_572 | htr1__HO_L_vs_NRC-1 |
| hal_575 | htlD__HO_D_vs_NRC-1 |
| hal_576 | htlD__HO_L_vs_NRC-1 |
| hal_577 | htlD__LO_D_vs_NRC-1 |
| hal_578 | htlD__LO_L_vs_NRC-1 |
| hal_579 | gap__HO_D_vs_NRC-1c |
| hal_58 | dark1__3360m_vs_NRC-1c |
| hal_580 | gap__HO_L_vs_NRC-1c |
| hal_581 | gap__LO_D_vs_NRC-1c |
| hal_582 | gap__LO_L_vs_NRC-1c |
| hal_59 | dark1__3600m_vs_NRC-1c |
| hal_592 | gamma__0000gy-0000m |
| hal_593 | gamma__0000gy-0010m |
| hal_594 | gamma__0000gy-0030m |
| hal_595 | gamma__0000gy-0060m |
| hal_597 | gamma__0000gy-0960m |
| hal_60 | dark1__3870m_vs_NRC-1c |
| hal_61 | dark1__4110m_vs_NRC-1c |
| hal_618 | bop__HO_D_vs_NRC-1 |
| hal_623 | boa4__HO_L_vs_NRC-1 |
| hal_625 | boa4__LO_L_vs_NRC-1 |
| hal_629 | boa1__LO_L_vs_NRC-1 |
| hal_631 | ark__HO_L_vs_NRC-1 |
| hal_633 | ark__LO_L_vs_NRC-1 |
| hal_634 | afsQ2__HO_D_vs_NRC-1 |
| hal_635 | afsQ2__HO_L_vs_NRC-1 |
| hal_636 | afsQ2__LO_D_vs_NRC-1 |
| hal_892 | gamma4__2500gy-060min |
| hal_895 | gamma__2500gy-0000m |
| hal_896 | gamma__2500gy-0010m |
| hal_897 | gamma__2500gy-0020m |
| hal_898 | gamma__2500gy-0030m |
| hal_899 | gamma__2500gy-0040m |
| hal_900 | gamma__2500gy-0060m |
| hal_901 | gamma__2500gy-0120m |
| hal_902 | gamma__2500gy-0240m |
| hal_903 | gamma__2500gy-0480m |
| hal_904 | gamma__2500gy-0960m |
| hal_964 | uv__200jm2_L_060m |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| hal_1208_1 | 0.0000 | 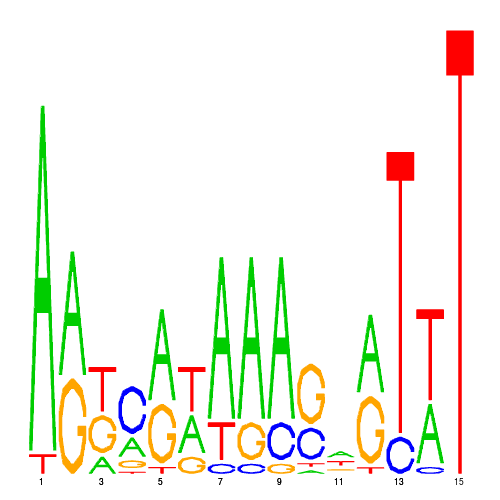 |
| hal_1208_2 | 5600.0000 | 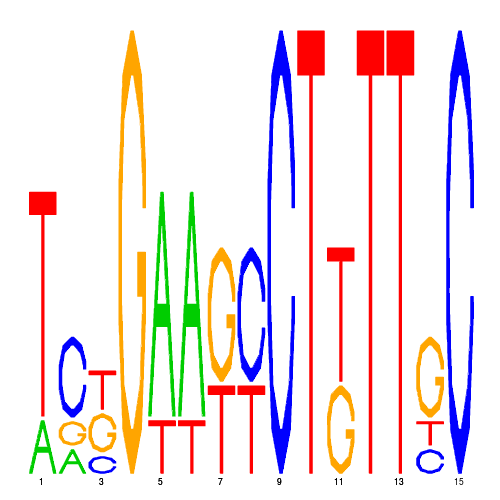 |
GREs assigned to hal_1208
| GRE ID | PSSM |
|---|---|
| hal_61 | 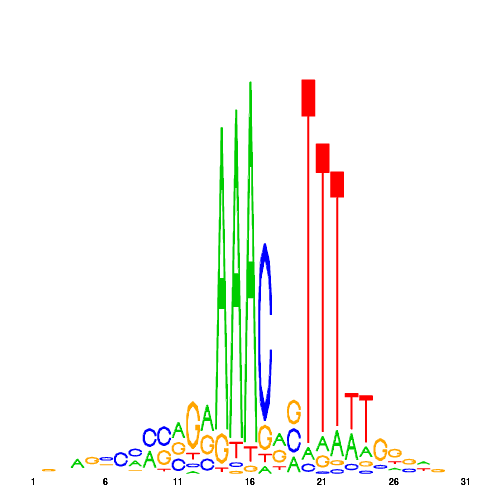 |
| hal_0 |  |
Corems associated with hal_1208
| Corem |
|---|
| hc2913 |
| hc36696 |
| hc80 |
| hc8054 |