hal_16964 Co-expression
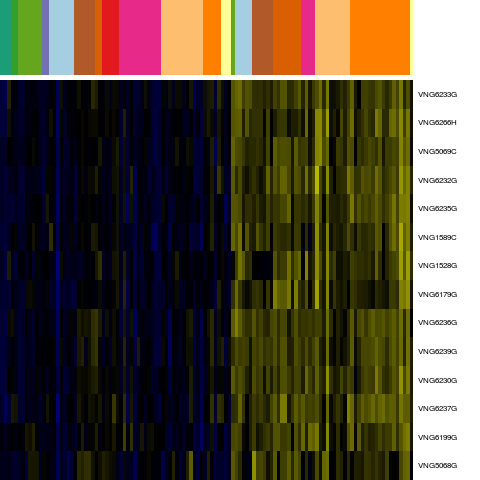
Genes in hal_16964
Conditions in hal_16964
| Condition ID | Condition Name |
|---|---|
| hal_1009 | idr1_20min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1011 | idr1_60min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1017 | sirR_20min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1018 | sirR_40min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1024 | ura3_0min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1025 | ura3_20min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1026 | ura3_40min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1027 | ura3_60min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1120 | gamma3__0000gy-0000m |
| hal_1121 | gamma3__0000gy-0010m |
| hal_1123 | gamma3__0000gy-0030m |
| hal_1124 | gamma3__0000gy-0040m |
| hal_1125 | gamma3__0000gy-0050m |
| hal_1237 | 20060717_Cmyc_rep2_0.223_vs_NRC-1e.sig |
| hal_1239 | 20060717_Cmyc_rep2_0.883_vs_NRC-1e.sig |
| hal_1252 | 20060308_KO_tfbD_rep1_0.424_vs_NRC-1e.sig |
| hal_1280 | Cmy_cspD2_2_hOD_vs_NRC-1d.sig |
| hal_1281 | Cmy_cspD2_2_mOD_vs_NRC-1d.sig |
| hal_1282 | Cmy_cspD1_1_hOD_vs_NRC-1d.sig |
| hal_1285 | Cmy_cspD1_2_mOD_vs_NRC-1d.sig |
| hal_1286 | 20050726_Cmyc_tfbF_rep2_0.180_vs_NRC-1d.sig |
| hal_1289 | 20050726_Cmyc_tfbF_rep2_0.918_vs_NRC-1d.sig |
| hal_1298 | 20050726_Cmyc_tfbC_rep1_0.132_vs_NRC-1d.sig |
| hal_1299 | 20050726_Cmyc_tfbC_rep1_0.394_vs_NRC-1d.sig |
| hal_1302 | 20050726_Cmyc_tfbB_rep2_0.152_vs_NRC-1d.sig |
| hal_1303 | 20050726_Cmyc_tfbB_rep2_0.496_vs_NRC-1d.sig |
| hal_1304 | 20050726_Cmyc_tfbB_rep2_0.840_vs_NRC-1d.sig |
| hal_1306 | 20050726_Cmyc_tfbB_rep1_0.157_vs_NRC-1d.sig |
| hal_1336 | 20050620_Cmyc_tfbD_rep2_0.153_vs_NRC-1d.sig |
| hal_1345 | 20050620_Cmyc_rep1_0.207_vs_NRC-1d.sig |
| hal_1346 | 20050620_Cmyc_rep1_0.474_vs_NRC-1d.sig |
| hal_1347 | 20050620_Cmyc_rep1_1.052_vs_NRC-1d.sig |
| hal_1349 | H2O2_Recov._set_1_0mM_000min_vs_NRC-1d.sig |
| hal_1351 | H2O2_Recov._set_1_0mM_020min_vs_NRC-1d.sig |
| hal_1353 | H2O2_Recov._set_1_0mM_040min_vs_NRC-1d.sig |
| hal_1354 | H2O2_Recov._set_1_0mM_050min_vs_NRC-1d.sig |
| hal_1356 | H2O2_Recov._set_1_0mM_120min_vs_NRC-1d.sig |
| hal_1360 | KO_asnC.2_1.3133_vs_NRC-1_h1.sig |
| hal_138 | circadian_dark3_cycling_1500min_vs_NRC-1d.sig |
| hal_142 | circadian_dark3_cycling_2160min_vs_NRC-1d.sig |
| hal_143 | circadian_dark3_cycling_2220min_vs_NRC-1d.sig |
| hal_1447 | sDura3D1179_pZn_d0.005mM_t+030m_vs_NRC-1h1.sig |
| hal_1448 | sDura3D1179_pZn_d0.005mM_t+060m_vs_NRC-1h1.sig |
| hal_1449 | sDura3D1179_pZn_d0.005mM_t+180m_vs_NRC-1h1.sig |
| hal_146 | circadian_dark3_cycling_2700min_vs_NRC-1d.sig |
| hal_309 | ZnSO4_set2_010min_vs_NRC-1h1.sig |
| hal_313 | ZnSO4_set2_030min_vs_NRC-1h1.sig |
| hal_317 | ZnSO4_set2_120min_vs_NRC-1h1.sig |
| hal_318 | ZnSO4_set2_160min_vs_NRC-1h1.sig |
| hal_319 | ZnSO4_set2_320min_vs_NRC-1h1.sig |
| hal_408 | ura3_0a_vs_NRC-1d.sig |
| hal_409 | ura3_0b_vs_NRC-1d.sig |
| hal_410 | ura3_Cu_0700um_a_vs_NRC-1d.sig |
| hal_414 | ura3_Cu_0850um_b_vs_NRC-1d.sig |
| hal_416 | ura3_Mn_0800um_a_vs_NRC-1d.sig |
| hal_417 | ura3_Mn_1500um_a_vs_NRC-1d.sig |
| hal_419 | ura3_Mn_0800um_b_vs_NRC-1d.sig |
| hal_420 | ura3_Mn_1500um_b_vs_NRC-1d.sig |
| hal_434 | ni__0000um_vs_NRC-1c |
| hal_435 | ni__0500um_vs_NRC-1c |
| hal_437 | ni__1500um_vs_NRC-1c |
| hal_510 | ura3__LO_D_vs_NRC-1 |
| hal_511 | ura3__LO_L_vs_NRC-1 |
| hal_648 | NRC1_H2O_1860_0.758_rep1_vs_NRC-1g.sig |
| hal_649 | NRC1_H2O_2280_0.781_rep1_vs_NRC-1g.sig |
| hal_650 | NRC1_H2O_2760_0.913_rep1_vs_NRC-1g.sig |
| hal_651 | NRC1_H2O_3120_1.023_rep1_vs_NRC-1g.sig |
| hal_653 | NRC1_Uracil_0890_0.420_rep2_vs_NRC-1g.sig |
| hal_656 | NRC1_Uracil_2280_0.881_rep2_vs_NRC-1g.sig |
| hal_657 | NRC1_Uracil_2760_1.133_rep2_vs_NRC-1g.sig |
| hal_658 | NRC1_Uracil_3120_1.296_rep2_vs_NRC-1g.sig |
| hal_688 | NRC1_03960_0.409_rep2_vs_NRC-1g.sig |
| hal_691 | NRC1_05400_0.786_rep2_vs_NRC-1g.sig |
| hal_692 | NRC1_05778_0.722_rep2_vs_NRC-1g.sig |
| hal_693 | NRC1_07020_1.318_rep2_vs_NRC-1g.sig |
| hal_694 | NRC1_07740_2.681_rep2_vs_NRC-1g.sig |
| hal_695 | NRC1_08460_3.840_rep2_vs_NRC-1g.sig |
| hal_698 | NRC1_10980_4.935_rep2_vs_NRC-1g.sig |
| hal_699 | NRC1_11700_5.730_rep2_vs_NRC-1g.sig |
| hal_72 | circadian_drk8_cycling_0180min_vs_NRC-1e.sig |
| hal_743 | H2O2_Recov._set_2_0mM_020min_vs_NRC-1f.sig |
| hal_744 | H2O2_Recov._set_2_0mM_030min_vs_NRC-1f.sig |
| hal_745 | H2O2_Recov._set_2_0mM_040min_vs_NRC-1f.sig |
| hal_746 | H2O2_Recov._set_2_0mM_050min_vs_NRC-1f.sig |
| hal_747 | H2O2_Recov._set_2_0mM_060min_vs_NRC-1f.sig |
| hal_749 | H2O2_Recov._set_2_0mM_240min_vs_NRC-1f.sig |
| hal_76 | circadian_drk8_cycling_0900min_vs_NRC-1e.sig |
| hal_79 | circadian_drk8_cycling_1440min_vs_NRC-1e.sig |
| hal_794 | ura3_-glu_a_OD0.6_vs_NRC1e.sig |
| hal_798 | ura3_-glu_a_OD1.2_vs_NRC1e.sig |
| hal_799 | 1451_growth_t4_exp2_0.5OD_vs_NRC1f.sig |
| hal_80 | circadian_drk8_cycling_1620min_vs_NRC-1e.sig |
| hal_801 | 1451_growth_t2_exp3_0.2OD_vs_NRC1f.sig |
| hal_804 | ura3_growth_t4_exp2_1.2OD_vs_NRC1f.sig |
| hal_807 | ura3_growth_t3_exp3_0.8OD_vs_NRC1f.sig |
| hal_808 | ura3_growth_t4_exp3_1.2OD_vs_NRC1f.sig |
| hal_815 | 1451_-glu_ab_OD0.2_vs_NRC1e.sig |
| hal_816 | 1451_-glu_a_OD0.6repl_vs_NRC1e.sig |
| hal_817 | 1451_-glu_a_OD1.2repl_vs_NRC1g.sig |
| hal_82 | circadian_drk8_cycling_1980min_vs_NRC-1e.sig |
| hal_820 | ura3_+glu_a_OD0.2_vs_NRC1e.sig |
| hal_823 | ura3_+glu_b_OD0.2_vs_NRC1g.sig |
| hal_826 | ura3_-glu_a_OD0.2_vs_NRC1e.sig |
| hal_827 | ura3_-glu_a_OD0.6repl_vs_NRC1e.sig |
| hal_828 | ura3_-glu_a_OD1.2repl_vs_NRC1e.sig |
| hal_83 | circadian_drk8_cycling_2160min_vs_NRC-1e.sig |
| hal_864 | ura3_growth_t3exp1_0.8OD_vs_NRC1e.sig |
| hal_87 | circadian_drk8_cycling_2880min_vs_NRC-1e.sig |
| hal_89 | circadian_drk8_cycling_3240min_vs_NRC-1e.sig |
| hal_914 | H2O2_Const._set_2_25mM_000min_vs_NRC-1f.sig |
| hal_917 | H2O2_Const._set_2_25mM_120min_vs_NRC-1f.sig |
| hal_918 | H2O2_Const._set_2_25mM_240min_vs_NRC-1f.sig |
| hal_957 | PQ_Const._set_1_4mM_030min_vs_NRC-1f.sig |
| hal_958 | PQ_Const._set_1_4mM_060min_vs_NRC-1f.sig |
| hal_982 | ura3_20min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_983 | ura3_40min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_990 | idr1_20min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_992 | idr1_60min_1mM_EDTA_vs_NRC1-h1.sig |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| hal_16964_1 | 0.0000 | 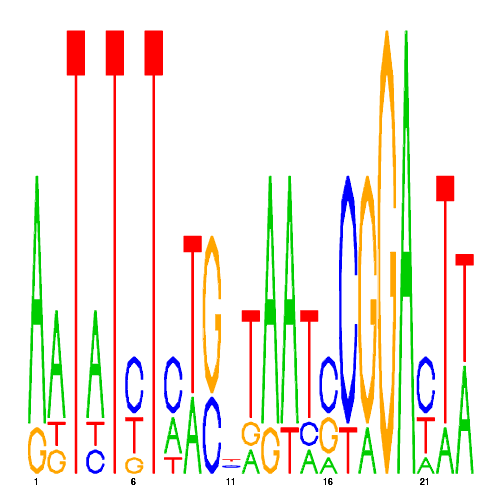 |
| hal_16964_2 | 0.0110 | 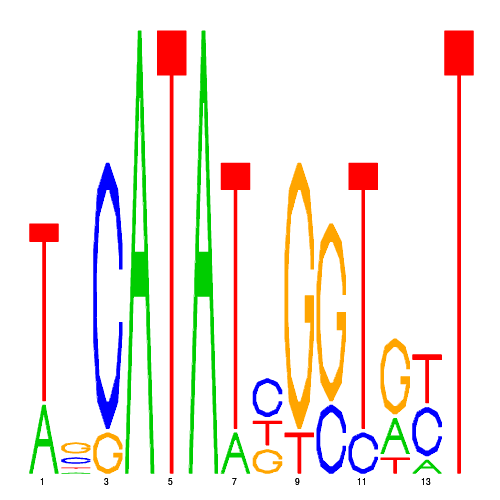 |
GREs assigned to hal_16964
| GRE ID | PSSM |
|---|---|
| hal_0 |  |
| hal_0 |  |
Corems associated with hal_16964
| Corem |
|---|
| hc39290 |
| hc39314 |
| hc39317 |
| hc39330 |