hal_73862 Co-expression
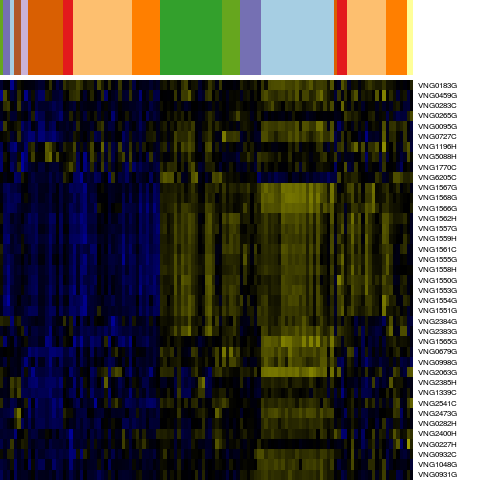
Genes in hal_73862
Conditions in hal_73862
| Condition ID | Condition Name |
|---|---|
| hal_1027 | ura3_60min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1032 | KO_trh6.2_0.1677_vs_NRC-1_h1.sig |
| hal_1033 | KO_trh6.2_0.3621_vs_NRC-1_h1.sig |
| hal_1035 | KO_trh6.2_0.838_vs_NRC-1_h1.sig |
| hal_1048 | KO_trh7.1_0.1442_vs_NRC-1_h1.sig |
| hal_1049 | KO_trh7.1_0.3293_vs_NRC-1_h1.sig |
| hal_1050 | KO_trh7.1_0.5217_vs_NRC-1_h1.sig |
| hal_1111 | o2_set3_0120_min_HO2_vs_NRC1f.sig |
| hal_1119 | o2_set3_0660_min_HO2_vs_NRC1f.sig |
| hal_1156 | 20061006_Cmyc_tfbD_rep1_0.185_vs_NRC-1e.sig |
| hal_1158 | 20061006_Cmyc_tfbD_rep1_0.899_vs_NRC-1e.sig |
| hal_1159 | 20061006_Cmyc_rep2_0.179_vs_NRC-1e.sig |
| hal_1160 | 20061006_Cmyc_rep2_0.399_vs_NRC-1e.sig |
| hal_1161 | 20061006_Cmyc_rep2_0.833_vs_NRC-1e.sig |
| hal_1180 | 20060818_KO_tfbC_rep1_0.188_vs_NRC-1e.sig |
| hal_1183 | 20060818_KO_tfbA_rep2_0.208_vs_NRC-1e.sig |
| hal_1195 | 20060818_KO_bat_rep2_0.183_vs_NRC-1e.sig |
| hal_1213 | 20060717_Cmyc_tbpD_rep2_0.185_vs_NRC-1d.sig |
| hal_1214 | 20060717_Cmyc_tbpD_rep2_0.413_vs_NRC-1d.sig |
| hal_1229 | 20060717_Cmyc_tbpB_rep1_0.431_vs_NRC-1e.sig |
| hal_1234 | 20060717_tbpA_Cmyc_rep1_0.199_vs_NRC-1e.sig |
| hal_1237 | 20060717_Cmyc_rep2_0.223_vs_NRC-1e.sig |
| hal_1238 | 20060717_Cmyc_rep2_0.407_vs_NRC-1e.sig |
| hal_1295 | 20050726_Cmyc_tfbC_rep2_0.430_vs_NRC-1d.sig |
| hal_1324 | 20050620_Cmyc_tfbG_rep2_0.170_vs_NRC-1d.sig |
| hal_1327 | 20050620_Cmyc_tfbG_rep1_0.180_vs_NRC-1d.sig |
| hal_1329 | 20050620_Cmyc_tfbG_rep1_1.080_vs_NRC-1d.sig |
| hal_1379 | NRC-1.2_0.3895_vs_NRC-1_h1.sig |
| hal_1380 | NRC-1.2_1.1065_vs_NRC-1_h1.sig |
| hal_1389 | KO_ura3.1_0.1857_vs_NRC-1_h1.sig |
| hal_1390 | KO_ura3.1_0.3501_vs_NRC-1_h1.sig |
| hal_1392 | KO_ura3.1_0.525_vs_NRC-1_h1.sig |
| hal_1398 | NRC-1_0.3279_vs_NRC-1_h1.sig |
| hal_1399 | NRC-1_0.3882_vs_NRC-1_h1.sig |
| hal_1400 | NRC-1_1.0338_vs_NRC-1_h1.sig |
| hal_1409 | gamma3__2500gy-0000m |
| hal_1414 | gamma3__2500gy-0050m |
| hal_1416 | gamma3__2500gy-0120m |
| hal_1417 | gamma3__2500gy-0240m |
| hal_1431 | sDura3D1179_pCu_d0.700mM_t+015m_vs_NRC-1h1.sig |
| hal_1432 | sDura3D1179_pCu_d0.700mM_t+030m_vs_NRC-1h1.sig |
| hal_1435 | sDura3D1179_pCu_d0.700mM_t-015m_vs_NRC-1h1.sig |
| hal_154 | circadian_dark3_ctrl_2160min_vs_NRC-1d.sig |
| hal_205 | Circadian_Dark9_Control_0000min_vs_NRC-1g.sig |
| hal_206 | Circadian_Dark9_Control_0180min_vs_NRC-1g.sig |
| hal_207 | Circadian_Dark9_Control_0360min_vs_NRC-1g.sig |
| hal_208 | Circadian_Dark9_Control_0535min_vs_NRC-1g.sig |
| hal_209 | Circadian_Dark9_Control_0720min_vs_NRC-1g.sig |
| hal_210 | Circadian_Dark9_Control_1080min_vs_NRC-1g.sig |
| hal_211 | Circadian_Dark9_Control_1265min_vs_NRC-1g.sig |
| hal_212 | Circadian_Dark9_Control_1440min_vs_NRC-1g.sig |
| hal_214 | Circadian_Dark9_Control_1800min_vs_NRC-1g.sig |
| hal_215 | Circadian_Dark9_Control_1980min_vs_NRC-1g.sig |
| hal_216 | Circadian_Dark9_Control_2160min_vs_NRC-1g.sig |
| hal_217 | Circadian_Dark9_Control_2340min_vs_NRC-1g.sig |
| hal_218 | Circadian_Dark9_Control_2520min_vs_NRC-1g.sig |
| hal_219 | Circadian_Dark9_Control_2700min_vs_NRC-1g.sig |
| hal_221 | Circadian_Dark9_Control_3060min_vs_NRC-1g.sig |
| hal_222 | Circadian_Dark9_Control_3240min_vs_NRC-1g.sig |
| hal_224 | Circadian_Dark9_Control_3600min_vs_NRC-1g.sig |
| hal_225 | Circadian_Dark9_Control_3780min_vs_NRC-1g.sig |
| hal_227 | Circadian_Dark9_Control_4140min_vs_NRC-1g.sig |
| hal_228 | Circadian_Dark9_Control_4320min_vs_NRC-1g.sig |
| hal_229 | Circadian_Dark9_Control_4500min_vs_NRC-1g.sig |
| hal_291 | Cu_005_vs_NRC-1e.sig |
| hal_292 | Cu_010_vs_NRC-1e.sig |
| hal_293 | Cu_020_vs_NRC-1e.sig |
| hal_294 | Cu_040_vs_NRC-1e.sig |
| hal_295 | Cu_080_vs_NRC-1e.sig |
| hal_297 | Cu_320_vs_NRC-1e.sig |
| hal_298 | ZnSO4_ts_set-1_-005min_vs_NRC-1h1.sig |
| hal_299 | ZnSO4_ts_set-1_000min_vs_NRC-1h1.sig |
| hal_301 | ZnSO4_ts_set-1_010min_vs_NRC-1h1.sig |
| hal_303 | ZnSO4_ts_set-1_040min_vs_NRC-1h1.sig |
| hal_304 | ZnSO4_ts_set-1_080min_vs_NRC-1h1.sig |
| hal_305 | ZnSO4_ts_set-1_160min_vs_NRC-1h1.sig |
| hal_337 | Cu_vng1179_KO_set1_000min_vs_NRC-1h1.sig |
| hal_338 | Cu_vng1179_KO_set1_005min_vs_NRC-1h1.sig |
| hal_339 | Cu_vng1179_KO_set1_010min_vs_NRC-1h1.sig |
| hal_340 | Cu_vng1179_KO_set1_020min_vs_NRC-1h1.sig |
| hal_341 | Cu_vng1179_KO_set1_040min_vs_NRC-1h1.sig |
| hal_342 | Cu_vng1179_KO_set1_080min_vs_NRC-1h1.sig |
| hal_343 | Cu_vng1179_KO_set1_160min_vs_NRC-1h1.sig |
| hal_408 | ura3_0a_vs_NRC-1d.sig |
| hal_409 | ura3_0b_vs_NRC-1d.sig |
| hal_410 | ura3_Cu_0700um_a_vs_NRC-1d.sig |
| hal_414 | ura3_Cu_0850um_b_vs_NRC-1d.sig |
| hal_419 | ura3_Mn_0800um_b_vs_NRC-1d.sig |
| hal_421 | ura3_Mn_1000um_b_vs_NRC-1d.sig |
| hal_446 | idr2_60min_+Fe_vs_NRC1-h1.sig |
| hal_457 | ura3_40min_+Fe_vs_NRC1-h1.sig |
| hal_487 | zim__HO_D_vs_NRC-1 |
| hal_490 | zim__LO_L_vs_NRC-1 |
| hal_510 | ura3__LO_D_vs_NRC-1 |
| hal_511 | ura3__LO_L_vs_NRC-1 |
| hal_513 | tfb_D_vs_NRC-1c |
| hal_514 | tfb_C_vs_NRC-1c |
| hal_515 | tfb_B_vs_NRC-1c |
| hal_516 | tfb_A_vs_NRC-1c |
| hal_551 | NRC-1__HO_D_vs_NRC-1 |
| hal_559 | kinA2_I__HO_D_vs_NRC-1 |
| hal_598 | EMS_Treated__000uM-000 |
| hal_603 | EMS_Treated__000uM-050 |
| hal_616 | NRC-1_vs_NRC-1c.sig |
| hal_769 | MDM200gSalt_0.199_vs_NRC-1e.sig |
| hal_770 | MDM200gSalt_0.388_vs_NRC-1e.sig |
| hal_771 | MDM200gSalt_0.466_vs_NRC-1e.sig |
| hal_790 | ura3_growth_t2_exp2_0.5OD_vs_NRC1f.sig |
| hal_793 | ura3_+glu_a_OD0.6_vs_NRC1e.sig |
| hal_794 | ura3_-glu_a_OD0.6_vs_NRC1e.sig |
| hal_797 | ura3_+glu_a_OD1.2_vs_NRC1e.sig |
| hal_798 | ura3_-glu_a_OD1.2_vs_NRC1e.sig |
| hal_805 | ura3_growth_t1_exp3_0.3OD_vs_NRC1f.sig |
| hal_806 | ura3_growth_t2_exp3_0.5OD_vs_NRC1f.sig |
| hal_821 | ura3_+glu_a_OD0.6repl_vs_NRC1e.sig |
| hal_822 | ura3_+glu_a_OD1.2repl_vs_NRC1e.sig |
| hal_828 | ura3_-glu_a_OD1.2repl_vs_NRC1e.sig |
| hal_830 | ura3_-glu_b_OD0.6repl_vs_NRC1g.sig |
| hal_981 | ura3_0min_1mM_EDTA_vs_NRC1-h1.sig |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| hal_73862_1 | 0.0000 | 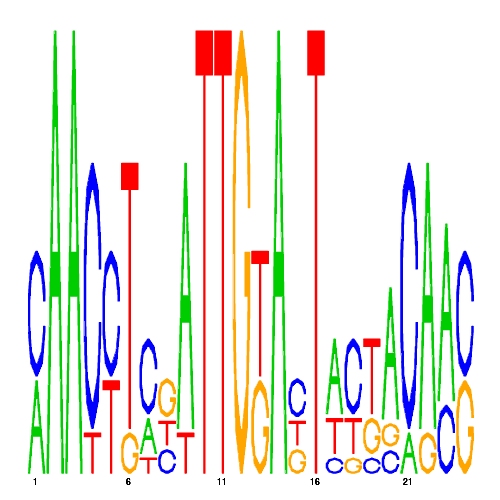 |
| hal_73862_2 | 0.0002 | 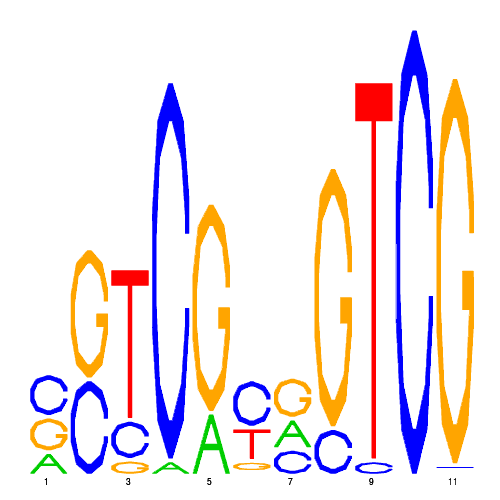 |
GREs assigned to hal_73862
| GRE ID | PSSM |
|---|---|
| hal_170 |  |
| hal_0 |  |
Corems associated with hal_73862
| Corem |
|---|
| hc27703 |
| hc8493 |