hal_9108 Co-expression
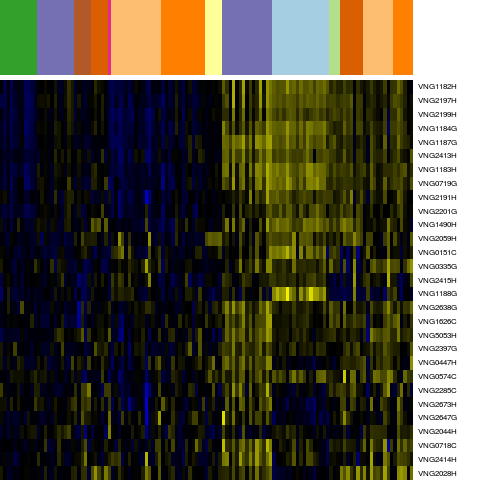
Genes in hal_9108
Conditions in hal_9108
| Condition ID | Condition Name |
|---|---|
| hal_1024 | ura3_0min_-EDTA_CM_vs_NRC1-h1.sig |
| hal_1032 | KO_trh6.2_0.1677_vs_NRC-1_h1.sig |
| hal_1035 | KO_trh6.2_0.838_vs_NRC-1_h1.sig |
| hal_1092 | o2_set2_L_005_vs_NRC-1d.sig |
| hal_1093 | o2_set2_L_010_vs_NRC-1d.sig |
| hal_1097 | o2_set2_L_090_vs_NRC-1d.sig |
| hal_1098 | o2_set2_L_135_vs_NRC-1d.sig |
| hal_1100 | o2_set2_L_270_vs_NRC-1d.sig |
| hal_1104 | o2_set3_0005_min_HO2_vs_NRC1f.sig |
| hal_1106 | o2_set3_0020_min_HO2_vs_NRC1f.sig |
| hal_1107 | o2_set3_0030_min_HO2_vs_NRC1f.sig |
| hal_1108 | o2_set3_0045_min_HO2_vs_NRC1f.sig |
| hal_1109 | o2_set3_0060_min_HO2_vs_NRC1f.sig |
| hal_1110 | o2_set3_0090_min_HO2_vs_NRC1f.sig |
| hal_1111 | o2_set3_0120_min_HO2_vs_NRC1f.sig |
| hal_1112 | o2_set3_0180_min_HO2_vs_NRC1f.sig |
| hal_1113 | o2_set3_0240_min_HO2_vs_NRC1f.sig |
| hal_1116 | o2_set3_0420_min_HO2_vs_NRC1f.sig |
| hal_1118 | o2_set3_0600_min_HO2_vs_NRC1f.sig |
| hal_1119 | o2_set3_0660_min_HO2_vs_NRC1f.sig |
| hal_1183 | 20060818_KO_tfbA_rep2_0.208_vs_NRC-1e.sig |
| hal_1213 | 20060717_Cmyc_tbpD_rep2_0.185_vs_NRC-1d.sig |
| hal_1251 | 20060308_KO_tfbD_rep1_0.190_vs_NRC-1e.sig |
| hal_1252 | 20060308_KO_tfbD_rep1_0.424_vs_NRC-1e.sig |
| hal_1266 | 20060308_KO_tbpC_rep2_0.181_vs_NRC-1e.sig |
| hal_1267 | 20060308_KO_tbpC_rep2_0.419_vs_NRC-1e.sig |
| hal_1278 | Cmy_cspD2_1_hOD_vs_NRC-1d.sig |
| hal_1279 | Cmy_cspD2_1_mOD_vs_NRC-1d.sig |
| hal_1280 | Cmy_cspD2_2_hOD_vs_NRC-1d.sig |
| hal_1281 | Cmy_cspD2_2_mOD_vs_NRC-1d.sig |
| hal_1282 | Cmy_cspD1_1_hOD_vs_NRC-1d.sig |
| hal_1283 | Cmy_cspD1_1_mOD_vs_NRC-1d.sig |
| hal_1285 | Cmy_cspD1_2_mOD_vs_NRC-1d.sig |
| hal_1319 | 20050620_NRC1_rep2_0.505_vs_NRC-1d.sig |
| hal_1320 | 20050620_NRC1_rep2_1.030_vs_NRC-1d.sig |
| hal_1331 | 20050620_Cmyc_tfbE_rep2_0.383_vs_NRC-1d.sig |
| hal_1348 | NRC-1e_vs_NRC-1h1.sig |
| hal_1380 | NRC-1.2_1.1065_vs_NRC-1_h1.sig |
| hal_1397 | NRC-1_0.1779_vs_NRC-1_h1.sig |
| hal_1419 | Dun_Supernat._005minb_vs_NRC-1g.sig |
| hal_1422 | Dun_Supernat._040minb_vs_NRC-1g.sig |
| hal_1423 | Dun_Supernat._080minb_vs_NRC-1g.sig |
| hal_1479 | sDura3_pNULL_d0.000mM_t+180m_vs_NRC-1h1.sig |
| hal_1480 | sDura3_pNULL_d0.000mM_t-015m_vs_NRC-1h1.sig |
| hal_357 | Cu_vng149_KO_set1_040min_vs_NRC-1h1.sig |
| hal_368 | Cu_vng700_KO_set2_-030min_vs_NRC-1h1.sig |
| hal_372 | Cu_vng700_KO_set2_020min_vs_NRC-1h1.sig |
| hal_374 | Cu_vng700_KO_set2_080min_vs_NRC-1h1.sig |
| hal_375 | Cu_vng700_KO_set2_160min_vs_NRC-1h1.sig |
| hal_408 | ura3_0a_vs_NRC-1d.sig |
| hal_409 | ura3_0b_vs_NRC-1d.sig |
| hal_410 | ura3_Cu_0700um_a_vs_NRC-1d.sig |
| hal_411 | ura3_Cu_0850um_a_vs_NRC-1d.sig |
| hal_412 | ura3_Cu_1000um_a_vs_NRC-1d.sig |
| hal_413 | ura3_Cu_0700um_b_vs_NRC-1d.sig |
| hal_414 | ura3_Cu_0850um_b_vs_NRC-1d.sig |
| hal_415 | ura3_Cu_1000um_b_vs_NRC-1d.sig |
| hal_416 | ura3_Mn_0800um_a_vs_NRC-1d.sig |
| hal_417 | ura3_Mn_1500um_a_vs_NRC-1d.sig |
| hal_418 | ura3_Mn_1000um_a_vs_NRC-1d.sig |
| hal_419 | ura3_Mn_0800um_b_vs_NRC-1d.sig |
| hal_420 | ura3_Mn_1500um_b_vs_NRC-1d.sig |
| hal_504 | VNG0019H__HO_L_vs_NRC-1c |
| hal_506 | VNG0019H__LO_L_vs_NRC-1c |
| hal_509 | ura3__HO_L_vs_NRC-1 |
| hal_551 | NRC-1__HO_D_vs_NRC-1 |
| hal_552 | NRC-1__HO_L_vs_NRC-1 |
| hal_554 | NRC-1__LO_L_vs_NRC-1 |
| hal_562 | kinA2_I__LO_L_vs_NRC-1 |
| hal_577 | htlD__LO_D_vs_NRC-1 |
| hal_616 | NRC-1_vs_NRC-1c.sig |
| hal_617 | NRC-1c_vs_NRC-1d.sig |
| hal_635 | afsQ2__HO_L_vs_NRC-1 |
| hal_686 | NRC-1f_vs_NRC-1g.sig |
| hal_71 | circadian_drk8_cycling_0000min_vs_NRC-1e.sig |
| hal_73 | circadian_drk8_cycling_0360min_vs_NRC-1e.sig |
| hal_74 | circadian_drk8_cycling_0540min_vs_NRC-1e.sig |
| hal_75 | circadian_drk8_cycling_0720min_vs_NRC-1e.sig |
| hal_76 | circadian_drk8_cycling_0900min_vs_NRC-1e.sig |
| hal_77 | circadian_drk8_cycling_1080min_vs_NRC-1e.sig |
| hal_78 | circadian_drk8_cycling_1260min_vs_NRC-1e.sig |
| hal_786 | 1451_growth_t1_exp2_0.1OD_vs_NRC1f.sig |
| hal_787 | 1451_growth_t2_exp2_0.2OD_vs_NRC1f.sig |
| hal_788 | 1451_growth_t3_exp2_0.3OD_vs_NRC1f.sig |
| hal_789 | ura3_growth_t1_exp2_0.3OD_vs_NRC1f.sig |
| hal_79 | circadian_drk8_cycling_1440min_vs_NRC-1e.sig |
| hal_795 | 1451_+glu_a_OD0.6_vs_NRC1e.sig |
| hal_797 | ura3_+glu_a_OD1.2_vs_NRC1e.sig |
| hal_798 | ura3_-glu_a_OD1.2_vs_NRC1e.sig |
| hal_799 | 1451_growth_t4_exp2_0.5OD_vs_NRC1f.sig |
| hal_80 | circadian_drk8_cycling_1620min_vs_NRC-1e.sig |
| hal_801 | 1451_growth_t2_exp3_0.2OD_vs_NRC1f.sig |
| hal_802 | 1451_growth_t3_exp3_0.3OD_vs_NRC1f.sig |
| hal_803 | 1451_growth_t4_exp3_0.6OD_vs_NRC1f.sig |
| hal_804 | ura3_growth_t4_exp2_1.2OD_vs_NRC1f.sig |
| hal_806 | ura3_growth_t2_exp3_0.5OD_vs_NRC1f.sig |
| hal_81 | circadian_drk8_cycling_1800min_vs_NRC-1e.sig |
| hal_811 | 1451_+glu_a_OD1.2repl_vs_NRC1e.sig |
| hal_813 | 1451_+glu_b_OD0.6repl_vs_NRC1g.sig |
| hal_814 | 1451_+glu_b_OD1.2repl_vs_NRC1g.sig |
| hal_816 | 1451_-glu_a_OD0.6repl_vs_NRC1e.sig |
| hal_817 | 1451_-glu_a_OD1.2repl_vs_NRC1g.sig |
| hal_818 | 1451_-glu_b_OD0.6repl_vs_NRC1g.sig |
| hal_819 | 1451_-glu_b_OD1.2repl_vs_NRC1g.sig |
| hal_82 | circadian_drk8_cycling_1980min_vs_NRC-1e.sig |
| hal_823 | ura3_+glu_b_OD0.2_vs_NRC1g.sig |
| hal_830 | ura3_-glu_b_OD0.6repl_vs_NRC1g.sig |
| hal_84 | circadian_drk8_cycling_2340min_vs_NRC-1e.sig |
| hal_848 | PQ_Const._set_3_4mM_160min_vs_NRC-1h1.sig |
| hal_85 | circadian_drk8_cycling_2520min_vs_NRC-1e.sig |
| hal_856 | KO_ura3.2_0.1716_vs_NRC-1_h1.sig |
| hal_857 | KO_ura3.2_0.3207_vs_NRC-1_h1.sig |
| hal_859 | KO_ura3.2_0.5084_vs_NRC-1_h1.sig |
| hal_86 | circadian_drk8_cycling_2700min_vs_NRC-1e.sig |
| hal_860 | 1451_growth_t1exp1_0.2OD_vs_NRC1e.sig |
| hal_861 | 1451_growth_t2exp1_0.3OD_vs_NRC1e.sig |
| hal_87 | circadian_drk8_cycling_2880min_vs_NRC-1e.sig |
| hal_88 | circadian_drk8_cycling_3060min_vs_NRC-1e.sig |
| hal_89 | circadian_drk8_cycling_3240min_vs_NRC-1e.sig |
| hal_983 | ura3_40min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_984 | ura3_60min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_986 | idr1-2_20min_1mM_EDTA_vs_NRC1-h1.sig |
| hal_987 | idr1-2_40min_1mM_EDTA_vs_NRC1-h1.sig |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| hal_9108_1 | 0.0000 | 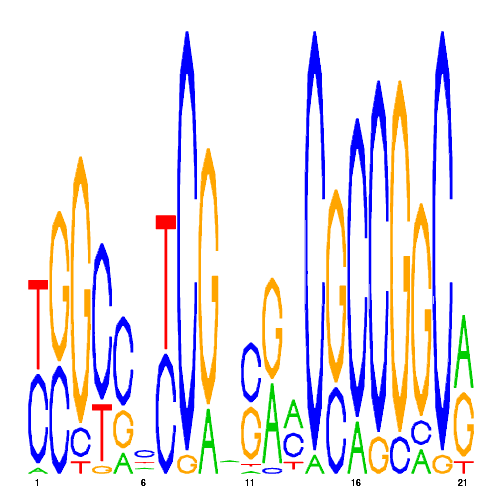 |
| hal_9108_2 | 40.0000 | 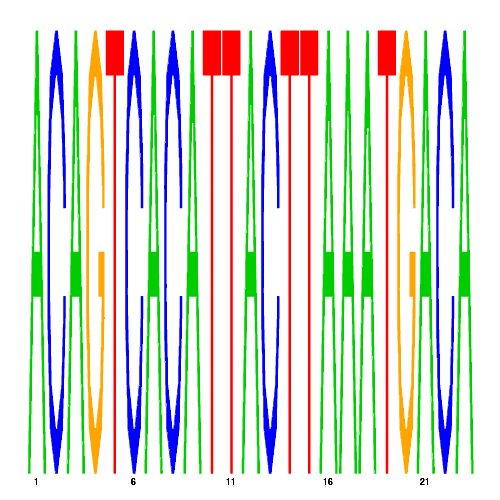 |
GREs assigned to hal_9108
| GRE ID | PSSM |
|---|---|
| hal_0 |  |
| hal_128 |  |
Corems associated with hal_9108
| Corem |
|---|
| hc171 |