Regulator CueR
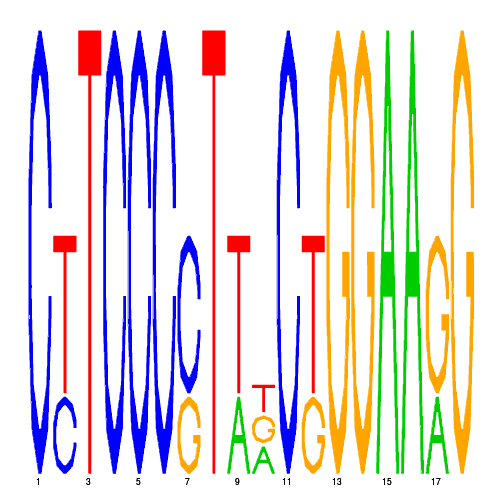
GREs
| GRE ID | PSSM | Score |
|---|---|---|
| eco_58 | 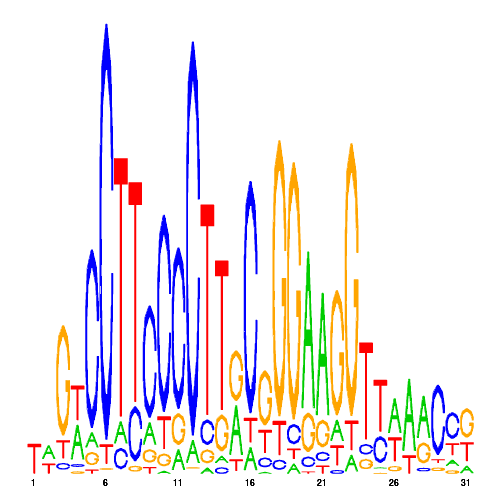 |
0.0000000000000000 |
Corems
| Corem ID | GRE | pval |
|---|---|---|
| ec519242 | eco_58 | 0.0003291516000 |
| ec519253 | eco_58 | 0.0003291516000 |
| ec519588 | eco_58 | 0.0002671351000 |
| ec519598 | eco_58 | 0.0000041102490 |
| ec519609 | eco_58 | 0.0000082005690 |
| ec520496 | eco_58 | 0.0001691290000 |
| ec526846 | eco_58 | 0.0004371302000 |
| ec527232 | eco_58 | 0.0003468782000 |
| ec528728 | eco_58 | 0.0006047243000 |
| ec557434 | eco_58 | 0.0002914201000 |
Genes
| Gene name | Common name | Description | Start | Stop | Strand | Chromosome | GRE | pval |
|---|---|---|---|---|---|---|---|---|
| b0069 | sgrR | DNA-binding transcriptional regulator | 77299 | 75644 | - | NC_000913.2 | eco_58 | 0.0001028898000 |
| b0071 | leuD | isopropylmalate isomerase small subun | 79453 | 78848 | - | NC_000913.2 | eco_58 | 0.0000000167183 |
| b0122 | yacC | orf, hypothetical protein | 136917 | 136570 | - | NC_000913.2 | eco_58 | 0.0000000000000 |
| b0123 | cueO | multicopper oxidase (laccase) | 137083 | 138633 | + | NC_000913.2 | eco_58 | 0.0000000000000 |
| b0130 | yadE | predicted polysaccharide deacetylase lipoprotein | 145081 | 146310 | + | NC_000913.2 | eco_58 | 0.0000005264981 |
| b0158 | btuF | vitamin B12-transporter protein BtuF | 178462 | 177662 | - | NC_000913.2 | eco_58 | 0.0000000000001 |
| b0160 | dgt | deoxyguanosinetriphosphate triphosphohydrolase | 179237 | 180754 | + | NC_000913.2 | eco_58 | 0.0000000015232 |
| b0484 | copA | copper transporter | 510603 | 508099 | - | NC_000913.2 | eco_58 | 0.0000000000000 |
| b0592 | fepB | iron-enterobactin transporter subun | 623733 | 622777 | - | NC_000913.2 | eco_58 | 0.0004106278000 |
| b0772 | ybhC | predicted pectinesterase | 806504 | 805221 | - | NC_000913.2 | eco_58 | 0.0000021991410 |
| b0781 | moaA | molybdenum cofactor biosynthesis protein A | 816267 | 817256 | + | NC_000913.2 | eco_58 | 0.0000001287339 |
| b0801 | ybiC | predicted dehydrogenase | 835574 | 836659 | + | NC_000913.2 | eco_58 | 0.0000009475886 |
| b0815 | ybiP | predicted hydrolase, inner membrane | 851820 | 850237 | - | NC_000913.2 | eco_58 | 0.0000436230100 |
| b0869 | ybjT | putative dTDP-glucose enzyme | 907505 | 906075 | - | NC_000913.2 | eco_58 | 0.0000229214100 |
| b0874 | ybjE | putative surface protein | 914080 | 913181 | - | NC_000913.2 | eco_58 | 0.0000000000063 |
| b0931 | pncB | nicotinate phosphoribosyltransferase | 989579 | 988377 | - | NC_000913.2 | eco_58 | 0.0000051694340 |
| b1420 | mokB | regulatory peptide | 1490153 | 1489986 | - | NC_000913.2 | eco_58 | 0.0000601352500 |
| b1634 | tppB | putative tripeptide transporter permease | 1710793 | 1712295 | + | NC_000913.2 | eco_58 | 0.0000103147500 |
| b1693 | aroD | 3-dehydroquinate dehydratase | 1772710 | 1773468 | + | NC_000913.2 | eco_58 | 0.0000020581940 |
| b1867 | yecD | orf, hypothetical protein | 1948856 | 1949422 | + | NC_000913.2 | eco_58 | 0.0000051694340 |
| b2152 | yeiB | conserved inner membrane protein | 2240989 | 2239832 | - | NC_000913.2 | eco_58 | 0.0000021080860 |
| b2290 | yfbQ | aspartate aminotransferase | 2405583 | 2406800 | + | NC_000913.2 | eco_58 | 0.0000355129100 |
| b2369 | evgA | DNA-binding response regulator in two-component regulatory system with EvgS | 2481777 | 2482391 | + | NC_000913.2 | eco_58 | 0.0000141963600 |
| b2599 | pheA | fused chorismate mutase P/prephenate dehydratase | 2735767 | 2736927 | + | NC_000913.2 | eco_58 | 0.0005450381000 |
| b3105 | yhaJ | predicted DNA-binding transcriptional regulator | 3252236 | 3251340 | - | NC_000913.2 | eco_58 | 0.0000378048900 |
| b3995 | rsd | stationary phase protein, binds sigma 70 RNA polymerase subun | 4194831 | 4194355 | - | NC_000913.2 | eco_58 | 0.0001489272000 |
| b3996 | nudC | NADH pyrophosphatase | 4194926 | 4195699 | + | NC_000913.2 | eco_58 | 0.0000418345000 |
Conditions
| Condition ID | Condition Name | GRE | pval |
|---|---|---|---|
| eco_102 | GSM118620 | 53 | 0.0118790300000 |
| eco_107 | GSM137283 | 53 | 0.0242073700000 |
| eco_109 | GSM1379 | 53 | 0.0494606900000 |
| eco_11 | Clean_ANA12 | 53 | 0.0156655800000 |
| eco_119 | GSM18262 | 53 | 0.0359861000000 |
| eco_12 | Clean_ANA13 | 53 | 0.0186531100000 |
| eco_122 | GSM18273 | 53 | 0.0132717600000 |
| eco_128 | GSM30821 | 53 | 0.0015830140000 |
| eco_13 | Clean_ANA6 | 53 | 0.0149462200000 |
| eco_134 | GSM37077 | 53 | 0.0090992990000 |
| eco_147 | GSM65630 | 53 | 0.0060930880000 |
| eco_149 | GSM67647 | 53 | 0.0414501900000 |
| eco_156 | GSM73140 | 53 | 0.0000033867300 |
| eco_157 | GSM73141 | 53 | 0.0077069880000 |
| eco_16 | Clean_DIAUX12 | 53 | 0.0280523500000 |
| eco_162 | GSM73152 | 53 | 0.0102229800000 |
| eco_163 | GSM73155 | 53 | 0.0048100880000 |
| eco_164 | GSM73157 | 53 | 0.0268326300000 |
| eco_165 | GSM73158 | 53 | 0.0353660600000 |
| eco_166 | GSM73159 | 53 | 0.0239905300000 |
| eco_167 | GSM73160 | 53 | 0.0026714400000 |
| eco_168 | GSM73257 | 53 | 0.0085747600000 |
| eco_171 | GSM73264 | 53 | 0.0001618647000 |
| eco_172 | GSM73265 | 53 | 0.0068556130000 |
| eco_173 | GSM73270 | 53 | 0.0300154800000 |
| eco_175 | GSM73272 | 53 | 0.0088612230000 |
| eco_176 | GSM73276 | 53 | 0.0073130160000 |
| eco_177 | GSM73277 | 53 | 0.0253764800000 |
| eco_181 | GSM77341 | 53 | 0.0014641250000 |
| eco_188 | GSM8116 | 53 | 0.0358976000000 |
| eco_19 | Clean_DIAUX7 | 53 | 0.0333453900000 |
| eco_193 | GSM82548 | 53 | 0.0336556100000 |
| eco_2 | 1285 | 53 | 0.0423573200000 |
| eco_206 | GSM83078 | 53 | 0.0117441500000 |
| eco_212 | GSM98723 | 53 | 0.0095845970000 |
| eco_213 | GSM98726 | 53 | 0.0394231400000 |
| eco_219 | GSM99103 | 53 | 0.0243110700000 |
| eco_232 | GSM99136 | 53 | 0.0443899200000 |
| eco_239 | GSM99173 | 53 | 0.0061493090000 |
| eco_249 | GSM99211 | 53 | 0.0188661800000 |
| eco_25 | Clean_TGC10 | 53 | 0.0092937330000 |
| eco_26 | Clean_TGC14 | 53 | 0.0319895700000 |
| eco_261 | 1595 | 53 | 0.0208701500000 |
| eco_262 | 1638 | 53 | 0.0204582500000 |
| eco_263 | 1639 | 53 | 0.0495181900000 |
| eco_266 | 5265 | 53 | 0.0346238700000 |
| eco_275 | Clean_H2O28 | 53 | 0.0007374332000 |
| eco_28 | Clean_TGC2 | 53 | 0.0247780400000 |
| eco_290 | EC_LAC_WILD_D | 53 | 0.0300343500000 |
| eco_293 | E-MEXP-244-raw-data-396455098.txt | 53 | 0.0444681400000 |
| eco_3 | 1593 | 53 | 0.0121803400000 |
| eco_300 | GADXW_exp8 | 53 | 0.0357915200000 |
| eco_301 | GADXW_exp9 | 53 | 0.0082421510000 |
| eco_31 | EC_GLY_GLY1_20_C | 53 | 0.0023668560000 |
| eco_329 | GSM1374 | 53 | 0.0337683000000 |
| eco_336 | GSM18275 | 53 | 0.0408325000000 |
| eco_34 | EC_GLY_GLY2_20_A | 53 | 0.0011491040000 |
| eco_347 | GSM65624 | 53 | 0.0137048500000 |
| eco_352 | GSM73134 | 53 | 0.0090321350000 |
| eco_354 | GSM73147 | 53 | 0.0138732500000 |
| eco_357 | GSM73262 | 53 | 0.0317223100000 |
| eco_359 | GSM73269 | 53 | 0.0286644300000 |
| eco_37 | EC_GLY_GLYA_44_B | 53 | 0.0070611250000 |
| eco_371 | GSM82552 | 53 | 0.0418457500000 |
| eco_372 | GSM82553 | 53 | 0.0267210800000 |
| eco_381 | GSM83081 | 53 | 0.0422946800000 |
| eco_386 | GSM98731 | 53 | 0.0482402400000 |
| eco_391 | GSM99105 | 53 | 0.0460192200000 |
| eco_406 | GSM99192 | 53 | 0.0033446720000 |
| eco_419 | 1637 | 53 | 0.0002193496000 |
| eco_423 | 9395 | 53 | 0.0032475270000 |
| eco_424 | Clean_ANA2 | 53 | 0.0162279900000 |
| eco_425 | Clean_TGC12 | 53 | 0.0095596250000 |
| eco_426 | Clean_TGC16 | 53 | 0.0066492830000 |
| eco_427 | Clean_TGC6 | 53 | 0.0396382300000 |
| eco_428 | Clean_TGC8 | 53 | 0.0005905193000 |
| eco_43 | EC_GLY_WILD_D | 53 | 0.0327594400000 |
| eco_430 | EC_GLY_GLY1_20_A | 53 | 0.0001215820000 |
| eco_436 | EC_LAC_LE_20_A | 53 | 0.0289306400000 |
| eco_438 | fruR | 53 | 0.0175648300000 |
| eco_439 | GADXW_exp1 | 53 | 0.0000321273700 |
| eco_440 | GADXW_exp11 | 53 | 0.0398994900000 |
| eco_441 | GADXW_exp14 | 53 | 0.0223171356000 |
| eco_442 | GADXW_exp15 | 53 | 0.0020605240000 |
| eco_455 | GSM118617 | 53 | 0.0350787400000 |
| eco_457 | GSM1377 | 53 | 0.0054171820000 |
| eco_459 | GSM18237 | 53 | 0.0187876200000 |
| eco_461 | GSM18284 | 53 | 0.0274212200000 |
| eco_463 | GSM30819 | 53 | 0.0365917000000 |
| eco_467 | GSM37074 | 53 | 0.0016619320000 |
| eco_474 | GSM65594 | 53 | 0.0458098800000 |
| eco_483 | GSM73136 | 53 | 0.0470221700000 |
| eco_484 | GSM73154 | 53 | 0.0002211882000 |
| eco_489 | GSM73282 | 53 | 0.0257236700000 |
| eco_499 | GSM82621 | 53 | 0.0411214100000 |
| eco_502 | GSM98720 | 53 | 0.0049341080000 |
| eco_510 | GSM99102 | 53 | 0.0385885000000 |
| eco_511 | GSM99104 | 53 | 0.0356392500000 |
| eco_514 | GSM99135 | 53 | 0.0363719700000 |
| eco_523 | GSM99389 | 53 | 0.0494997900000 |
| eco_525 | HEATSHOCK_CH | 53 | 0.0027185230000 |
| eco_528 | wt__G | 53 | 0.0479131900000 |
| eco_530 | 14830 | 53 | 0.0003827318000 |
| eco_531 | 1585 | 53 | 0.0309226800000 |
| eco_537 | Clean_TGC5 | 53 | 0.0153099100000 |
| eco_543 | EC_GLY_GLY1_20_B | 53 | 0.0421880100000 |
| eco_553 | EC_LAC_LC_20_B | 53 | 0.0430641400000 |
| eco_558 | E-MEXP-267-raw-data-495080223.txt | 53 | 0.0150238400000 |
| eco_563 | GSM101778 | 53 | 0.0155739100000 |
| eco_567 | GSM111423 | 53 | 0.0108335000000 |
| eco_57 | EC_LAC_WILD_E | 53 | 0.0107651500000 |
| eco_575 | GSM18253 | 53 | 0.0331530700000 |
| eco_58 | EC_LAC_WILD_F | 53 | 0.0459337800000 |
| eco_581 | GSM62197 | 53 | 0.0101434900000 |
| eco_583 | GSM63894 | 53 | 0.0144936800000 |
| eco_587 | GSM77345 | 53 | 0.0386835300000 |
| eco_590 | GSM77361 | 53 | 0.0483112300000 |
| eco_593 | GSM83061 | 53 | 0.0270105800000 |
| eco_607 | GSM99141 | 53 | 0.0295745600000 |
| eco_614 | GSM99210 | 53 | 0.0000059428490 |
| eco_617 | 1641 | 53 | 0.0064518730000 |
| eco_618 | 1642 | 53 | 0.0085022310000 |
| eco_621 | Clean_DIAUX15 | 53 | 0.0472517000000 |
| eco_626 | EC_GLY_GLY2_20_B | 53 | 0.0007397617000 |
| eco_627 | EC_GLY_GLY2_44_B | 53 | 0.0087075890000 |
| eco_63 | E-MEXP-244-raw-data-396455082.txt | 53 | 0.0051793280000 |
| eco_633 | EC_GLY_WILD_E | 53 | 0.0013376650000 |
| eco_640 | EC_LAC_LE_60_B | 53 | 0.0359200500000 |
| eco_641 | E-MEXP-244-raw-data-396455050.txt | 53 | 0.0045706210000 |
| eco_642 | E-MEXP-267-raw-data-495080335.txt | 53 | 0.0147063700000 |
| eco_643 | GSM101766 | 53 | 0.0323320300000 |
| eco_645 | GSM106338 | 53 | 0.0232525100000 |
| eco_649 | GSM118610 | 53 | 0.0179941600000 |
| eco_658 | GSM30823 | 53 | 0.0122676900000 |
| eco_66 | E-MEXP-245-raw-data-396455381.txt | 53 | 0.0406549300000 |
| eco_660 | GSM37070 | 53 | 0.0159154900000 |
| eco_663 | GSM65598 | 53 | 0.0026559830000 |
| eco_669 | GSM67665 | 53 | 0.0334302600000 |
| eco_67 | E-MEXP-267-raw-data-495080207.txt | 53 | 0.0071584780000 |
| eco_670 | GSM73126 | 53 | 0.0126507800000 |
| eco_673 | GSM73256 | 53 | 0.0038160080000 |
| eco_675 | GSM73274 | 53 | 0.0152933900000 |
| eco_676 | GSM73275 | 53 | 0.0057861130000 |
| eco_677 | GSM73280 | 53 | 0.0005381507000 |
| eco_678 | GSM77356 | 53 | 0.0154631300000 |
| eco_687 | GSM88917 | 53 | 0.0033696470000 |
| eco_688 | GSM89487 | 53 | 0.0414395600000 |
| eco_70 | GADXW_exp13 | 53 | 0.0038413180000 |
| eco_704 | GSM99380 | 53 | 0.0121068600000 |
| eco_707 | Clean_ANA15 | 53 | 0.0174443200000 |
| eco_708 | Clean_ANA9 | 53 | 0.0319447200000 |
| eco_71 | GADXW_exp6 | 53 | 0.0156391200000 |
| eco_717 | GADXW_exp4 | 53 | 0.0036657020000 |
| eco_726 | GSM63888 | 53 | 0.0113337800000 |
| eco_743 | GSM98722 | 53 | 0.0065246990000 |
| eco_752 | GSM99182 | 53 | 0.0256790300000 |
| eco_753 | 5281 | 53 | 0.0019047740000 |
| eco_754 | Clean_ANA17 | 53 | 0.0001659196000 |
| eco_762 | GADXW_exp10 | 53 | 0.0441472200000 |
| eco_763 | GADXW_exp5 | 53 | 0.0334723900000 |
| eco_766 | GSM116031 | 53 | 0.0067968870000 |
| eco_771 | GSM73127 | 53 | 0.0179532200000 |
| eco_776 | GSM77342 | 53 | 0.0487852400000 |
| eco_784 | crp__G | 53 | 0.0186285700000 |
| eco_788 | GSM101238 | 53 | 0.0455703800000 |
| eco_794 | GSM118601 | 53 | 0.0461597100000 |
| eco_797 | GSM73150 | 53 | 0.0228520100000 |
| eco_800 | GSM73284 | 53 | 0.0154210500000 |
| eco_81 | GSM101773 | 53 | 0.0105386000000 |
| eco_811 | Clean_DIAUX17 | 53 | 0.0146150700000 |
| eco_839 | GSM1370 | 53 | 0.0227593100000 |
| eco_842 | GSM65639 | 53 | 0.0298609800000 |
| eco_847 | GSM73135 | 53 | 0.0099109680000 |
| eco_856 | Clean_ANA20 | 53 | 0.0078087270000 |
| eco_858 | GSM118609 | 53 | 0.0137254200000 |
| eco_867 | GSM83075 | 53 | 0.0298812200000 |
| eco_87 | GSM111415 | 53 | 0.0397475800000 |
| eco_9 | 5287 | 53 | 0.0201849700000 |
| eco_98 | GSM118605 | 53 | 0.0324308900000 |