Regulator IscR
GREs
| GRE ID | PSSM | Score |
|---|---|---|
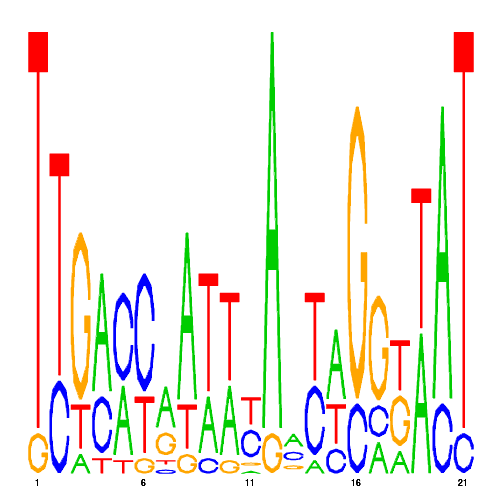
| eco_169 | 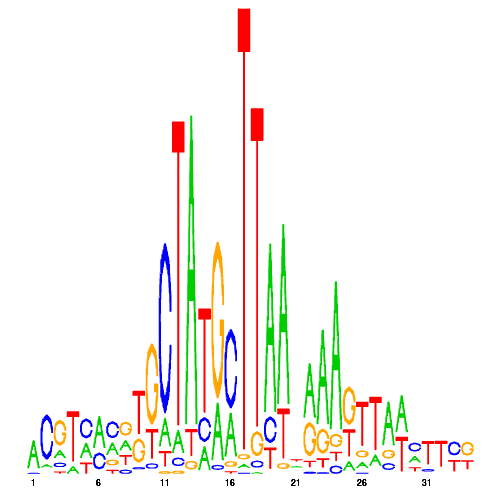 |
0.0072162349950057 |
| eco_185 |  |
0.0000000000000000 |
Corems
| Corem ID | GRE | pval |
|---|---|---|
| ec519630 | eco_169 | 0.0000000000225 |
| ec524775 | eco_169 | 0.0000412159100 |
| ec528619 | eco_169 | 0.0000000013598 |
| ec528622 | eco_169 | 0.0000228764600 |
| ec528626 | eco_169 | 0.0000000094325 |
| ec528631 | eco_169 | 0.0000002448303 |
| ec529321 | eco_169 | 0.0000000000000 |
| ec535079 | eco_169 | 0.0000000000000 |
| ec536158 | eco_169 | 0.0000264381000 |
| ec538659 | eco_169 | 0.0000016109070 |
| ec552920 | eco_185 | 0.0000362387800 |
Genes
| Gene name | Common name | Description | Start | Stop | Strand | Chromosome | GRE | pval |
|---|---|---|---|---|---|---|---|---|
| b0186 | ldcC | lysine decarboxylase 2, constitutive | 209679 | 211820 | + | NC_000913.2 | eco_169 | 0.0000000000000 |
| b0581 | ybdK | gamma-glutamyl:cysteine ligase | 606606 | 605488 | - | NC_000913.2 | eco_169 | 0.0000115363400 |
| b0677 | nagA | N-acetylglucosamine-6-phosphate deacetylase | 701974 | 700826 | - | NC_000913.2 | eco_169 | 0.0000002852839 |
| b0756 | galM | galactose-1-epimerase (mutarotase) | 788060 | 787020 | - | NC_000913.2 | eco_169 | 0.0000040210290 |
| b0839 | dacC | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 6a) | 879950 | 881152 | + | NC_000913.2 | eco_169 | 0.0000000002470 |
| b1103 | ycfF | orf, hypothetical protein | 1161108 | 1161467 | + | NC_000913.2 | eco_169 | 0.0000003229526 |
| b1327 | ycjY | predicted hydrolase | 1389877 | 1388957 | - | NC_000913.2 | eco_169 | 0.0000000007382 |
| b1341 | b1341 | orf, hypothetical protein | 1405819 | 1404587 | - | NC_000913.2 | eco_169 | 0.0000022413440 |
| b1342 | ydaN | zinc transporter | 1406074 | 1407057 | + | NC_000913.2 | eco_169 | 0.0000006414196 |
| b1399 | paaX | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive | 1461563 | 1462513 | + | NC_000913.2 | eco_169 | 0.0000000000000 |
| b1400 | paaY | predicted hexapeptide repeat acetyltransferase | 1462495 | 1463085 | + | NC_000913.2 | eco_169 | 0.0000004072895 |
| b1414 | ydcF | hypothetical protein | 1485259 | 1486059 | + | NC_000913.2 | eco_169 | 0.0000001069815 |
| b1428 | ydcK | predicted enzyme | 1498473 | 1497493 | - | NC_000913.2 | eco_169 | 0.0000001017387 |
| b1450 | b1450 | orf, hypothetical protein | 1518286 | 1518951 | + | NC_000913.2 | eco_169 | 0.0000000071004 |
| b1585 | ynfC | orf, hypothetical protein | 1655481 | 1654771 | - | NC_000913.2 | eco_169 | 0.0000000000000 |
| b1586 | b1586 | orf, hypothetical protein | 1655589 | 1655894 | + | NC_000913.2 | eco_169 | 0.0000000000000 |
| b1614 | ydgA | hypothetical protein | 1687876 | 1689384 | + | NC_000913.2 | eco_169 | 0.0000000000000 |
| b1664 | ydhQ | hypothetical protein | 1744151 | 1742895 | - | NC_000913.2 | eco_169 | 0.0000002796558 |
| b1675 | ydhZ | hypothetical protein | 1753165 | 1752956 | - | NC_000913.2 | eco_169 | 0.0000012110720 |
| b1679 | sufE | cysteine desufuration protein SufE | 1757314 | 1756898 | - | NC_000913.2 | eco_169 | 0.0000000000003 |
| b1703 | ydiA | hypothetical protein | 1785469 | 1786302 | + | NC_000913.2 | eco_169 | 0.0000000009748 |
| b1710 | btuE | predicted glutathione peroxidase | 1792133 | 1791582 | - | NC_000913.2 | eco_169 | 0.0000054661020 |
| b1927 | amyA | cytoplasmic alpha-amylase | 2004180 | 2005667 | + | NC_000913.2 | eco_169 | 0.0000007180410 |
| b1953 | b1953 | orf, hypothetical protein | 2023010 | 2023237 | + | NC_000913.2 | eco_169 | 0.0001464724000 |
| b1990 | erfK | conserved protein with NAD(P)-binding Rossmann-fold domain | 2061347 | 2060415 | - | NC_000913.2 | eco_169 | 0.0000000000053 |
| b2301 | yfcF | predicted enzyme | 2418507 | 2417863 | - | NC_000913.2 | eco_169 | 0.0000017222040 |
| b2602 | yfiL | orf, hypothetical protein | 2739382 | 2739747 | + | NC_000913.2 | eco_169 | 0.0000000607969 |
| b2665 | ygaU | hypothetical protein | 2794808 | 2794359 | - | NC_000913.2 | eco_169 | 0.0000281298900 |
| b2666 | yqaE | predicted membrane protein | 2795050 | 2794892 | - | NC_000913.2 | eco_169 | 0.0000235519500 |
| b3100 | yqjK | hypothetical protein | 3248099 | 3248398 | + | NC_000913.2 | eco_169 | 0.0000000000000 |
| b3234 | degQ | serine endoprotease, periplasmic | 3378765 | 3380132 | + | NC_000913.2 | eco_169 | 0.0000003148128 |
| b3291 | mscL | large-conductance mechanosensitive channe | 3436046 | 3436456 | + | NC_000913.2 | eco_169 | 0.0000015403850 |
| b3474 | yhhT | orf, hypothetical protein | 3609888 | 3610937 | + | NC_000913.2 | eco_169 | 0.0000103756700 |
| b4127 | yjdJ | predicted acyltransferase with acyl-CoA N-acyltransferase domain | 4350108 | 4350380 | + | NC_000913.2 | eco_169 | 0.0000031363080 |
| b4214 | cysQ | PAPS (adenosine 3'-phosphate 5'-phosphosulfate) 3'(2'),5'-bisphosphate nucleotidase | 4434778 | 4435518 | + | NC_000913.2 | eco_169 | 0.0000000000450 |
| b0357 | yaiN | putative alpha helix chain | 379105 | 378830 | - | NC_000913.2 | eco_185 | 0.0000002935606 |
| b0993 | torS | sensor protein torS (regulator TorR) | 1055401 | 1052657 | - | NC_000913.2 | eco_185 | 0.0000000509395 |
| b0994 | torT | periplasmic sensory protein associated with the TorRS two-component regulatory system | 1055484 | 1056512 | + | NC_000913.2 | eco_185 | 0.0000000575981 |
| b1771 | ydjG | predicted oxidoreductase | 1853995 | 1853015 | - | NC_000913.2 | eco_185 | 0.0000063363240 |
| b2629 | yfjM | CP4-57 prophage; predicted protein | 2763798 | 2763535 | - | NC_000913.2 | eco_185 | 0.0000000000000 |
| b2630 | yfjN | CP4-57 prophage; RNase LS | 2763940 | 2765013 | + | NC_000913.2 | eco_185 | 0.0000000000000 |
| b2631 | yfjO | orf, hypothetical protein | 2765006 | 2765377 | + | NC_000913.2 | eco_185 | 0.0000000000000 |
| b3954 | yijO | predicted DNA-binding transcriptional regulator | 4146340 | 4145489 | - | NC_000913.2 | eco_185 | 0.0001810723000 |
Conditions
| Condition ID | Condition Name | GRE | pval |
|---|---|---|---|
| eco_10 | 8455 | 146 | 0.0026532790000 |
| eco_101 | GSM118615 | 160 | 0.0312267400000 |
| eco_101 | GSM118615 | 146 | 0.0352336500000 |
| eco_102 | GSM118620 | 160 | 0.0133818400000 |
| eco_104 | GSM120192 | 160 | 0.0325240600000 |
| eco_105 | GSM120193 | 146 | 0.0293284000000 |
| eco_106 | GSM1371 | 160 | 0.0422321000000 |
| eco_107 | GSM137283 | 160 | 0.0057936470000 |
| eco_11 | Clean_ANA12 | 160 | 0.0076981150000 |
| eco_110 | GSM1380 | 146 | 0.0482826200000 |
| eco_111 | GSM18235 | 160 | 0.0118286900000 |
| eco_114 | GSM18252 | 160 | 0.0357286400000 |
| eco_115 | GSM18255 | 146 | 0.0493560200000 |
| eco_116 | GSM18256 | 160 | 0.0000109677300 |
| eco_118 | GSM18260 | 160 | 0.0037238090000 |
| eco_119 | GSM18262 | 160 | 0.0300826600000 |
| eco_123 | GSM18279 | 146 | 0.0162046600000 |
| eco_124 | GSM18285 | 146 | 0.0131518900000 |
| eco_125 | GSM30815 | 160 | 0.0083688950000 |
| eco_127 | GSM30820 | 160 | 0.0086838400000 |
| eco_131 | GSM37069 | 146 | 0.0044548550000 |
| eco_133 | GSM37075 | 146 | 0.0003097155000 |
| eco_134 | GSM37077 | 146 | 0.0010976410000 |
| eco_135 | GSM37886 | 160 | 0.0418009300000 |
| eco_136 | GSM38547 | 146 | 0.0458451500000 |
| eco_137 | GSM62194 | 146 | 0.0149826600000 |
| eco_141 | GSM63889 | 160 | 0.0300674000000 |
| eco_145 | GSM65626 | 160 | 0.0396250800000 |
| eco_147 | GSM65630 | 146 | 0.0342141400000 |
| eco_15 | Clean_DIAUX11 | 160 | 0.0004824783000 |
| eco_15 | Clean_DIAUX11 | 146 | 0.0398985800000 |
| eco_153 | GSM73130 | 146 | 0.0475360600000 |
| eco_156 | GSM73140 | 146 | 0.0447316300000 |
| eco_161 | GSM73151 | 160 | 0.0402724200000 |
| eco_164 | GSM73157 | 146 | 0.0268326300000 |
| eco_165 | GSM73158 | 146 | 0.0036293320000 |
| eco_165 | GSM73158 | 160 | 0.0278863400000 |
| eco_169 | GSM73258 | 146 | 0.0058929030000 |
| eco_17 | Clean_DIAUX13 | 146 | 0.0473565600000 |
| eco_170 | GSM73263 | 160 | 0.0003827444000 |
| eco_170 | GSM73263 | 146 | 0.0365119100000 |
| eco_175 | GSM73272 | 160 | 0.0314914200000 |
| eco_177 | GSM73277 | 160 | 0.0432960100000 |
| eco_181 | GSM77341 | 160 | 0.0062990880000 |
| eco_183 | GSM77351 | 160 | 0.0127660100000 |
| eco_187 | GSM7886 | 146 | 0.0352006600000 |
| eco_190 | GSM8120 | 160 | 0.0077565880000 |
| eco_193 | GSM82548 | 146 | 0.0170558200000 |
| eco_194 | GSM82551 | 146 | 0.0387663100000 |
| eco_195 | GSM82603 | 160 | 0.0360316800000 |
| eco_195 | GSM82603 | 146 | 0.0403295500000 |
| eco_20 | Clean_H2O210 | 160 | 0.0311957300000 |
| eco_200 | GSM83060 | 146 | 0.0083603160000 |
| eco_204 | GSM83071 | 146 | 0.0337839700000 |
| eco_21 | Clean_H2O211 | 146 | 0.0103456300000 |
| eco_212 | GSM98723 | 146 | 0.0160961300000 |
| eco_216 | GSM99090 | 160 | 0.0007292568000 |
| eco_216 | GSM99090 | 146 | 0.0429420400000 |
| eco_22 | Clean_H2O22 | 160 | 0.0272468400000 |
| eco_227 | GSM99122 | 146 | 0.0161289900000 |
| eco_23 | Clean_H2O24 | 146 | 0.0349963400000 |
| eco_230 | GSM99132 | 146 | 0.0366229100000 |
| eco_234 | GSM99152 | 160 | 0.0166263700000 |
| eco_235 | GSM99159 | 146 | 0.0483638100000 |
| eco_235 | GSM99159 | 160 | 0.0120527600000 |
| eco_236 | GSM99160 | 160 | 0.0082432520000 |
| eco_237 | GSM99164 | 146 | 0.0023051730000 |
| eco_238 | GSM99167 | 160 | 0.0003046851000 |
| eco_241 | GSM99184 | 160 | 0.0445718700000 |
| eco_244 | GSM99191 | 146 | 0.0371855100000 |
| eco_247 | GSM99203 | 146 | 0.0300721200000 |
| eco_251 | GSM99384 | 146 | 0.0457702800000 |
| eco_253 | GSM99393 | 146 | 0.0148293400000 |
| eco_254 | GSM99395 | 146 | 0.0411185000000 |
| eco_256 | GSM99401 | 146 | 0.0109586700000 |
| eco_263 | 1639 | 146 | 0.0424987400000 |
| eco_272 | Clean_DIAUX9 | 146 | 0.0068305650000 |
| eco_273 | Clean_H2O23 | 160 | 0.0311119000000 |
| eco_276 | cysB__G | 146 | 0.0393042300000 |
| eco_28 | Clean_TGC2 | 160 | 0.0096776910000 |
| eco_28 | Clean_TGC2 | 146 | 0.0127971100000 |
| eco_283 | EC_GLY_GLYE_44_B | 146 | 0.0137453100000 |
| eco_288 | EC_LAC_LD_20_B | 160 | 0.0357562100000 |
| eco_289 | EC_LAC_LE_20_B | 146 | 0.0195281000000 |
| eco_290 | EC_LAC_WILD_D | 146 | 0.0490369600000 |
| eco_293 | E-MEXP-244-raw-data-396455098.txt | 146 | 0.0441669500000 |
| eco_295 | E-MEXP-267-raw-data-495080191.txt | 146 | 0.0248533800000 |
| eco_298 | fur | 160 | 0.0292721200000 |
| eco_299 | GADXW_exp7 | 160 | 0.0038124850000 |
| eco_299 | GADXW_exp7 | 146 | 0.0180293100000 |
| eco_31 | EC_GLY_GLY1_20_C | 160 | 0.0305307300000 |
| eco_31 | EC_GLY_GLY1_20_C | 146 | 0.0358774800000 |
| eco_312 | GSM106504 | 146 | 0.0260866000000 |
| eco_314 | GSM106759 | 146 | 0.0309213900000 |
| eco_321 | GSM118611 | 160 | 0.0121637900000 |
| eco_324 | GSM118619 | 146 | 0.0006817594000 |
| eco_337 | GSM18281 | 160 | 0.0009729770000 |
| eco_341 | GSM37064 | 146 | 0.0374330900000 |
| eco_343 | GSM38549 | 160 | 0.0458431100000 |
| eco_344 | GSM62196 | 146 | 0.0354110000000 |
| eco_35 | EC_GLY_GLYA_20_A | 160 | 0.0176824500000 |
| eco_351 | GSM73122 | 146 | 0.0239896200000 |
| eco_352 | GSM73134 | 146 | 0.0252316800000 |
| eco_355 | GSM73255 | 160 | 0.0293954100000 |
| eco_355 | GSM73255 | 146 | 0.0105682600000 |
| eco_357 | GSM73262 | 146 | 0.0317223100000 |
| eco_358 | GSM73266 | 146 | 0.0216500200000 |
| eco_36 | EC_GLY_GLYA_20_C | 160 | 0.0070507360000 |
| eco_361 | GSM73281 | 146 | 0.0004771304000 |
| eco_364 | GSM77360 | 146 | 0.0446819200000 |
| eco_365 | GSM8113 | 146 | 0.0372695800000 |
| eco_366 | GSM8115 | 146 | 0.0387128300000 |
| eco_373 | GSM82598 | 146 | 0.0001155233000 |
| eco_376 | GSM83053 | 160 | 0.0186519900000 |
| eco_377 | GSM83057 | 160 | 0.0011464920000 |
| eco_38 | EC_GLY_GLYA_44_C | 146 | 0.0133596600000 |
| eco_382 | GSM83083 | 160 | 0.0281887900000 |
| eco_383 | GSM88912 | 160 | 0.0285388300000 |
| eco_386 | GSM98731 | 160 | 0.0364830400000 |
| eco_387 | GSM99084 | 146 | 0.0255176000000 |
| eco_389 | GSM99098 | 146 | 0.0247227000000 |
| eco_391 | GSM99105 | 146 | 0.0426539500000 |
| eco_396 | GSM99143 | 146 | 0.0309990700000 |
| eco_396 | GSM99143 | 160 | 0.0257240600000 |
| eco_398 | GSM99148 | 160 | 0.0375248100000 |
| eco_4 | 1597 | 160 | 0.0114431900000 |
| eco_401 | GSM99162 | 160 | 0.0016358220000 |
| eco_402 | GSM99166 | 160 | 0.0454118800000 |
| eco_407 | GSM99193 | 146 | 0.0195837800000 |
| eco_409 | GSM99200 | 146 | 0.0020248340000 |
| eco_414 | GSM99398 | 146 | 0.0223413200000 |
| eco_415 | GSM99400 | 146 | 0.0282823600000 |
| eco_416 | 1290 | 146 | 0.0373492200000 |
| eco_417 | 14829 | 146 | 0.0249947700000 |
| eco_418 | 14831 | 146 | 0.0083862650000 |
| eco_419 | 1637 | 160 | 0.0199627600000 |
| eco_426 | Clean_TGC16 | 146 | 0.0192021500000 |
| eco_429 | crpcysB | 146 | 0.0001356020000 |
| eco_43 | EC_GLY_WILD_D | 160 | 0.0132725600000 |
| eco_430 | EC_GLY_GLY1_20_A | 160 | 0.0136855000000 |
| eco_439 | GADXW_exp1 | 160 | 0.0357781900000 |
| eco_439 | GADXW_exp1 | 146 | 0.0417158300000 |
| eco_44 | EC_LAC_L2_20_C | 160 | 0.0453766600000 |
| eco_440 | GADXW_exp11 | 146 | 0.0001747313000 |
| eco_440 | GADXW_exp11 | 160 | 0.0037177640000 |
| eco_442 | GADXW_exp15 | 146 | 0.0051761820000 |
| eco_444 | GSM101767 | 146 | 0.0007204476000 |
| eco_453 | GSM114385 | 160 | 0.0018502110000 |
| eco_461 | GSM18284 | 160 | 0.0012616580000 |
| eco_463 | GSM30819 | 146 | 0.0365565000000 |
| eco_465 | GSM37068 | 146 | 0.0273291900000 |
| eco_467 | GSM37074 | 146 | 0.0304857600000 |
| eco_468 | GSM37885 | 146 | 0.0293638700000 |
| eco_475 | GSM65623 | 146 | 0.0273056600000 |
| eco_476 | GSM65628 | 160 | 0.0317781100000 |
| eco_479 | GSM67652 | 160 | 0.0172476400000 |
| eco_481 | GSM73119 | 146 | 0.0400453500000 |
| eco_483 | GSM73136 | 160 | 0.0360468100000 |
| eco_484 | GSM73154 | 146 | 0.0123896200000 |
| eco_485 | GSM73156 | 160 | 0.0081516300000 |
| eco_485 | GSM73156 | 146 | 0.0110209100000 |
| eco_487 | GSM73268 | 160 | 0.0198430900000 |
| eco_488 | GSM73279 | 146 | 0.0491318600000 |
| eco_489 | GSM73282 | 146 | 0.0471840500000 |
| eco_497 | GSM82605 | 146 | 0.0118005700000 |
| eco_497 | GSM82605 | 160 | 0.0087983670000 |
| eco_498 | GSM82610 | 160 | 0.0192347700000 |
| eco_500 | GSM88913 | 160 | 0.0444958900000 |
| eco_504 | GSM98728 | 146 | 0.0373850200000 |
| eco_506 | GSM99086 | 146 | 0.0261051200000 |
| eco_507 | GSM99088 | 160 | 0.0163243100000 |
| eco_510 | GSM99102 | 146 | 0.0379573200000 |
| eco_512 | GSM99131 | 160 | 0.0039467260000 |
| eco_512 | GSM99131 | 146 | 0.0051092390000 |
| eco_519 | GSM99194 | 146 | 0.0299645400000 |
| eco_521 | GSM99199 | 146 | 0.0403985700000 |
| eco_524 | GSM99394 | 146 | 0.0222846700000 |
| eco_527 | wt | 146 | 0.0003456821000 |
| eco_528 | wt__G | 160 | 0.0049021920000 |
| eco_532 | 1915 | 146 | 0.0134706200000 |
| eco_533 | 1916 | 160 | 0.0060916540000 |
| eco_533 | 1916 | 146 | 0.0282190900000 |
| eco_534 | Clean_DIAUX10 | 146 | 0.0425788100000 |
| eco_535 | Clean_DIAUX5 | 160 | 0.0308050500000 |
| eco_536 | Clean_H2O25 | 146 | 0.0129771000000 |
| eco_539 | crpcysB__G | 146 | 0.0185005200000 |
| eco_539 | crpcysB__G | 160 | 0.0442896800000 |
| eco_540 | crpfruR__G | 146 | 0.0096639300000 |
| eco_542 | EC_GLU_WILD_B | 160 | 0.0239697900000 |
| eco_544 | EC_GLY_GLY2_44_A | 160 | 0.0227941000000 |
| eco_545 | EC_GLY_GLYD_44_A | 160 | 0.0485446600000 |
| eco_546 | EC_GLY_GLYD_44_B | 160 | 0.0239437000000 |
| eco_55 | EC_LAC_LE_60_A | 146 | 0.0059242630000 |
| eco_558 | E-MEXP-267-raw-data-495080223.txt | 146 | 0.0115306500000 |
| eco_56 | EC_LAC_LE_60_C | 146 | 0.0024376500000 |
| eco_564 | GSM101780 | 146 | 0.0174380900000 |
| eco_567 | GSM111423 | 160 | 0.0411818600000 |
| eco_57 | EC_LAC_WILD_E | 146 | 0.0283122300000 |
| eco_572 | GSM1368 | 146 | 0.0394616900000 |
| eco_573 | GSM137282 | 160 | 0.0290309200000 |
| eco_573 | GSM137282 | 146 | 0.0119219900000 |
| eco_576 | GSM18261 | 160 | 0.0052392720000 |
| eco_578 | GSM18288 | 160 | 0.0032942610000 |
| eco_58 | EC_LAC_WILD_F | 146 | 0.0071558820000 |
| eco_580 | GSM37887 | 146 | 0.0307618100000 |
| eco_582 | GSM63891 | 160 | 0.0075811920000 |
| eco_584 | GSM73121 | 146 | 0.0078473990000 |
| eco_588 | GSM77349 | 146 | 0.0175135600000 |
| eco_589 | GSM77359 | 146 | 0.0054873400000 |
| eco_593 | GSM83061 | 160 | 0.0436178700000 |
| eco_600 | GSM98719 | 146 | 0.0112784100000 |
| eco_603 | GSM99087 | 146 | 0.0039506400000 |
| eco_604 | GSM99123 | 146 | 0.0204714400000 |
| eco_605 | GSM99130 | 146 | 0.0003399249000 |
| eco_606 | GSM99137 | 146 | 0.0248906800000 |
| eco_608 | GSM99151 | 160 | 0.0276394100000 |
| eco_614 | GSM99210 | 146 | 0.0053339920000 |
| eco_616 | 1278 | 146 | 0.0014463170000 |
| eco_620 | Clean_DIAUX14 | 146 | 0.0116951800000 |
| eco_621 | Clean_DIAUX15 | 160 | 0.0419405300000 |
| eco_622 | Clean_DIAUX16 | 146 | 0.0179816500000 |
| eco_623 | Clean_H2O21 | 146 | 0.0442199900000 |
| eco_624 | Clean_TGC1 | 160 | 0.0219982100000 |
| eco_627 | EC_GLY_GLY2_44_B | 146 | 0.0246338800000 |
| eco_633 | EC_GLY_WILD_E | 146 | 0.0102370000000 |
| eco_635 | EC_LAC_L3_20_B | 160 | 0.0111715700000 |
| eco_641 | E-MEXP-244-raw-data-396455050.txt | 160 | 0.0452966400000 |
| eco_642 | E-MEXP-267-raw-data-495080335.txt | 146 | 0.0305729600000 |
| eco_651 | GSM18236 | 146 | 0.0375377400000 |
| eco_653 | GSM18274 | 146 | 0.0112826400000 |
| eco_654 | GSM18276 | 160 | 0.0269068400000 |
| eco_656 | GSM18289 | 160 | 0.0183763200000 |
| eco_659 | GSM37066 | 146 | 0.0433038100000 |
| eco_662 | GSM63890 | 146 | 0.0170255000000 |
| eco_663 | GSM65598 | 160 | 0.0098516880000 |
| eco_669 | GSM67665 | 160 | 0.0277258300000 |
| eco_673 | GSM73256 | 146 | 0.0007394817000 |
| eco_68 | E-MEXP-267-raw-data-495080255.txt | 160 | 0.0234819300000 |
| eco_680 | GSM8114 | 160 | 0.0358136400000 |
| eco_681 | GSM82546 | 146 | 0.0066994270000 |
| eco_683 | GSM83063 | 146 | 0.0401569400000 |
| eco_685 | GSM83068 | 146 | 0.0026760300000 |
| eco_690 | GSM99082 | 146 | 0.0476794700000 |
| eco_692 | GSM99097 | 160 | 0.0209503700000 |
| eco_695 | GSM99115 | 146 | 0.0383935700000 |
| eco_697 | GSM99144 | 160 | 0.0185130800000 |
| eco_7 | 5268 | 160 | 0.0250791200000 |
| eco_700 | GSM99179 | 146 | 0.0269526200000 |
| eco_701 | GSM99190 | 146 | 0.0022220110000 |
| eco_706 | 1912 | 146 | 0.0404477800000 |
| eco_708 | Clean_ANA9 | 146 | 0.0319447200000 |
| eco_713 | EC_LAC_LD_20_C | 160 | 0.0205535600000 |
| eco_714 | EC_LAC_WILD_B | 160 | 0.0465551200000 |
| eco_716 | GADXW_exp2 | 146 | 0.0195008400000 |
| eco_717 | GADXW_exp4 | 160 | 0.0210141600000 |
| eco_719 | GSM120196 | 160 | 0.0453692100000 |
| eco_723 | GSM30822 | 160 | 0.0008899399000 |
| eco_724 | GSM35419 | 146 | 0.0327120900000 |
| eco_727 | GSM65589 | 146 | 0.0088714240000 |
| eco_731 | GSM67660 | 146 | 0.0119082900000 |
| eco_734 | GSM73252 | 146 | 0.0455532800000 |
| eco_736 | GSM77346 | 160 | 0.0006165966000 |
| eco_738 | GSM82544 | 160 | 0.0064153340000 |
| eco_739 | GSM83079 | 160 | 0.0013549940000 |
| eco_740 | GSM89485 | 146 | 0.0203768500000 |
| eco_743 | GSM98722 | 146 | 0.0379763400000 |
| eco_745 | GSM99095 | 146 | 0.0135261100000 |
| eco_746 | GSM99110 | 146 | 0.0274414200000 |
| eco_747 | GSM99120 | 146 | 0.0092228500000 |
| eco_747 | GSM99120 | 160 | 0.0264512500000 |
| eco_748 | GSM99163 | 160 | 0.0202977900000 |
| eco_749 | GSM99165 | 160 | 0.0121049300000 |
| eco_750 | GSM99178 | 160 | 0.0228112600000 |
| eco_753 | 5281 | 160 | 0.0142364900000 |
| eco_760 | EC_LAC_LD_60_B | 160 | 0.0274563000000 |
| eco_762 | GADXW_exp10 | 160 | 0.0192271700000 |
| eco_767 | GSM18254 | 146 | 0.0266930500000 |
| eco_769 | GSM18282 | 160 | 0.0497990700000 |
| eco_771 | GSM73127 | 146 | 0.0214033200000 |
| eco_773 | GSM73153 | 146 | 0.0075524220000 |
| eco_779 | GSM99127 | 146 | 0.0062708970000 |
| eco_782 | GSM99209 | 160 | 0.0095533360000 |
| eco_783 | Clean_DIAUX8 | 160 | 0.0475060500000 |
| eco_783 | Clean_DIAUX8 | 146 | 0.0180257700000 |
| eco_784 | crp__G | 146 | 0.0234483000000 |
| eco_787 | fruR__G | 146 | 0.0091418960000 |
| eco_788 | GSM101238 | 160 | 0.0178267300000 |
| eco_79 | GSM101771 | 160 | 0.0498667800000 |
| eco_79 | GSM101771 | 146 | 0.0054049110000 |
| eco_795 | GSM18246 | 146 | 0.0278186800000 |
| eco_797 | GSM73150 | 160 | 0.0111269800000 |
| eco_798 | GSM73253 | 146 | 0.0095322590000 |
| eco_799 | GSM73259 | 146 | 0.0404627300000 |
| eco_80 | GSM101772 | 160 | 0.0464985900000 |
| eco_80 | GSM101772 | 146 | 0.0059776170000 |
| eco_800 | GSM73284 | 160 | 0.0291340600000 |
| eco_801 | GSM82547 | 146 | 0.0240587700000 |
| eco_802 | GSM82620 | 160 | 0.0024111600000 |
| eco_803 | GSM98724 | 146 | 0.0407065700000 |
| eco_808 | GSM99638 | 160 | 0.0319036200000 |
| eco_81 | GSM101773 | 160 | 0.0115349400000 |
| eco_81 | GSM101773 | 146 | 0.0144312300000 |
| eco_815 | GSM101770 | 146 | 0.0298819400000 |
| eco_816 | GSM106755 | 146 | 0.0477379800000 |
| eco_817 | GSM18277 | 146 | 0.0385030000000 |
| eco_818 | GSM73129 | 146 | 0.0419978700000 |
| eco_819 | GSM77352 | 146 | 0.0011222780000 |
| eco_822 | GSM99154 | 146 | 0.0377310800000 |
| eco_826 | GSM106756 | 146 | 0.0404037700000 |
| eco_826 | GSM106756 | 160 | 0.0355381100000 |
| eco_83 | GSM106341 | 146 | 0.0434394300000 |
| eco_831 | GSM99139 | 146 | 0.0391087000000 |
| eco_833 | GSM99177 | 160 | 0.0081669120000 |
| eco_84 | GSM106757 | 146 | 0.0009453914000 |
| eco_84 | GSM106757 | 160 | 0.0006265342000 |
| eco_841 | GSM37076 | 146 | 0.0059881430000 |
| eco_846 | GSM18248 | 146 | 0.0295008500000 |
| eco_847 | GSM73135 | 146 | 0.0465179700000 |
| eco_851 | GSM99112 | 146 | 0.0112487900000 |
| eco_853 | GSM73128 | 146 | 0.0259344100000 |
| eco_854 | GSM99140 | 160 | 0.0479894800000 |
| eco_855 | GSM99161 | 160 | 0.0037487350000 |
| eco_857 | EC_LAC_LC_60_C | 160 | 0.0014670330000 |
| eco_857 | EC_LAC_LC_60_C | 146 | 0.0094111860000 |
| eco_859 | GSM18280 | 146 | 0.0294749100000 |
| eco_861 | GSM99124 | 146 | 0.0311081600000 |
| eco_861 | GSM99124 | 160 | 0.0288372500000 |
| eco_863 | GSM73118 | 146 | 0.0244012300000 |
| eco_864 | GSM99158 | 160 | 0.0161222700000 |
| eco_865 | Clean_DIAUX6 | 160 | 0.0446737600000 |
| eco_867 | GSM83075 | 160 | 0.0256074700000 |
| eco_868 | GSM73286 | 146 | 0.0044699360000 |
| eco_9 | 5287 | 146 | 0.0399856500000 |
| eco_92 | GSM114368 | 146 | 0.0327652000000 |
| eco_95 | GSM116024 | 160 | 0.0107295700000 |
| eco_99 | GSM118607 | 146 | 0.0188089300000 |