eco_2170 Co-expression
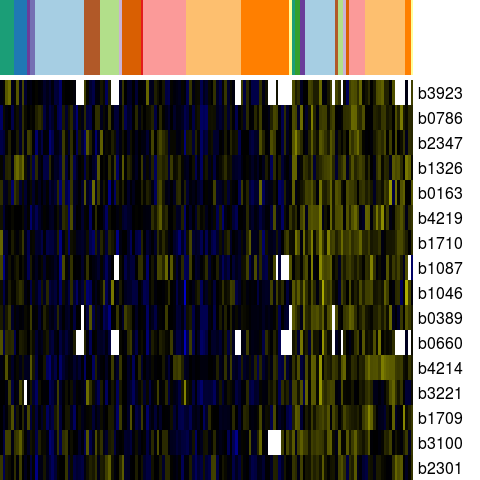
Genes in eco_2170
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0163 | yaeH | NP_414705.1 | hypothetical protein | 184095 | 183709 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0389 | yaiA | NP_414923.1 | hypothetical protein | 406203 | 406394 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0660 | ybeZ | NP_415193.2 | predicted protein with nucleoside triphosphate hydrolase domain | 692601 | 691561 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0786 | ybhL | NP_415307.1 | predicted inner membrane protein | 819107 | 819811 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1046 | ymdC | NP_415564.2 | putative synthase | 1105578 | 1106999 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1087 | yceF | NP_415605.2 | orf, hypothetical protein | 1145818 | 1145234 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1326 | mpaA | NP_415842.2 | murein peptide amidase A | 1388622 | 1387894 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1709 | btuD | NP_416224.1 | vitamin B12-transporter ATPase | 1791582 | 1790833 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1710 | btuE | NP_416225.1 | predicted glutathione peroxidase | 1792133 | 1791582 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2301 | yfcF | NP_416804.1 | predicted enzyme | 2418507 | 2417863 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2347 | yfdC | NP_416849.1 | predicted inner membrane protein | 2463323 | 2464255 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3100 | yqjK | NP_417571.1 | hypothetical protein | 3248099 | 3248398 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3221 | yhcH | NP_417688.1 | hypothetical protein | 3367500 | 3367036 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3923 | yiiT | NP_418358.1 | stress-induced protein | 4111316 | 4111744 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b4214 | cysQ | NP_418635.1 | PAPS (adenosine 3'-phosphate 5'-phosphosulfate) 3'(2'),5'-bisphosphate nucleotidase | 4434778 | 4435518 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b4219 | msrA | NP_418640.1 | methionine sulfoxide reductase A | 4440199 | 4439561 | - | NC_000913.2 |
Conditions in eco_2170
| Condition ID | Condition Name |
|---|---|
| eco_109 | GSM1379 |
| eco_116 | GSM18256 |
| eco_117 | GSM18257 |
| eco_119 | GSM18262 |
| eco_121 | GSM18264 |
| eco_123 | GSM18279 |
| eco_125 | GSM30815 |
| eco_135 | GSM37886 |
| eco_149 | GSM67647 |
| eco_15 | Clean_DIAUX11 |
| eco_154 | GSM73138 |
| eco_158 | GSM73144 |
| eco_163 | GSM73155 |
| eco_173 | GSM73270 |
| eco_175 | GSM73272 |
| eco_177 | GSM73277 |
| eco_182 | GSM77343 |
| eco_188 | GSM8116 |
| eco_19 | Clean_DIAUX7 |
| eco_200 | GSM83060 |
| eco_202 | GSM83065 |
| eco_204 | GSM83071 |
| eco_205 | GSM83077 |
| eco_21 | Clean_H2O211 |
| eco_210 | GSM98717 |
| eco_213 | GSM98726 |
| eco_220 | GSM99107 |
| eco_221 | GSM99108 |
| eco_226 | GSM99121 |
| eco_230 | GSM99132 |
| eco_232 | GSM99136 |
| eco_245 | GSM99196 |
| eco_246 | GSM99197 |
| eco_251 | GSM99384 |
| eco_258 | 1277 |
| eco_267 | 5278 |
| eco_268 | 5284 |
| eco_272 | Clean_DIAUX9 |
| eco_283 | EC_GLY_GLYE_44_B |
| eco_284 | EC_LAC_LA_20_B |
| eco_287 | EC_LAC_LB_20_C |
| eco_29 | crpfur |
| eco_294 | E-MEXP-245-raw-data-396455413.txt |
| eco_307 | GSM101775 |
| eco_308 | GSM101777 |
| eco_310 | GSM106339 |
| eco_32 | EC_GLY_GLY1_44_A |
| eco_320 | GSM116030 |
| eco_325 | GSM118621 |
| eco_33 | EC_GLY_GLY1_44_B |
| eco_331 | GSM1381 |
| eco_334 | GSM18266 |
| eco_340 | GSM23467 |
| eco_352 | GSM73134 |
| eco_353 | GSM73137 |
| eco_358 | GSM73266 |
| eco_369 | GSM82543 |
| eco_380 | GSM83074 |
| eco_385 | GSM98718 |
| eco_386 | GSM98731 |
| eco_391 | GSM99105 |
| eco_394 | GSM99125 |
| eco_396 | GSM99143 |
| eco_397 | GSM99145 |
| eco_405 | GSM99187 |
| eco_41 | EC_GLY_GLYC_20_C |
| eco_412 | GSM99390 |
| eco_416 | 1290 |
| eco_425 | Clean_TGC12 |
| eco_429 | crpcysB |
| eco_435 | EC_LAC_LC_20_C |
| eco_445 | GSM101768 |
| eco_446 | GSM101769 |
| eco_451 | GSM114371 |
| eco_458 | GSM1382 |
| eco_460 | GSM18259 |
| eco_468 | GSM37885 |
| eco_474 | GSM65594 |
| eco_475 | GSM65623 |
| eco_476 | GSM65628 |
| eco_478 | GSM67651 |
| eco_483 | GSM73136 |
| eco_484 | GSM73154 |
| eco_487 | GSM73268 |
| eco_488 | GSM73279 |
| eco_490 | GSM73290 |
| eco_508 | GSM99092 |
| eco_524 | GSM99394 |
| eco_529 | 1292 |
| eco_530 | 14830 |
| eco_538 | crp |
| eco_540 | crpfruR__G |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_548 | EC_GLY_GLYE_44_C |
| eco_553 | EC_LAC_LC_20_B |
| eco_556 | EC_LAC_LD_60_C |
| eco_562 | GADXW_exp12 |
| eco_57 | EC_LAC_WILD_E |
| eco_591 | GSM82608 |
| eco_592 | GSM83056 |
| eco_606 | GSM99137 |
| eco_608 | GSM99151 |
| eco_637 | EC_LAC_L3_60_C |
| eco_640 | EC_LAC_LE_60_B |
| eco_646 | GSM116027 |
| eco_653 | GSM18274 |
| eco_654 | GSM18276 |
| eco_665 | GSM65622 |
| eco_673 | GSM73256 |
| eco_681 | GSM82546 |
| eco_682 | GSM83054 |
| eco_689 | GSM98729 |
| eco_695 | GSM99115 |
| eco_701 | GSM99190 |
| eco_705 | GSM99397 |
| eco_709 | Clean_H2O29 |
| eco_721 | GSM18265 |
| eco_734 | GSM73252 |
| eco_736 | GSM77346 |
| eco_738 | GSM82544 |
| eco_741 | GSM89486 |
| eco_755 | Clean_DIAUX2 |
| eco_758 | EC_GLY_GLYD_44_C |
| eco_766 | GSM116031 |
| eco_767 | GSM18254 |
| eco_776 | GSM77342 |
| eco_777 | GSM83059 |
| eco_779 | GSM99127 |
| eco_783 | Clean_DIAUX8 |
| eco_785 | EC_LAC_L3_20_A |
| eco_793 | GSM116029 |
| eco_798 | GSM73253 |
| eco_799 | GSM73259 |
| eco_800 | GSM73284 |
| eco_804 | GSM99093 |
| eco_815 | GSM101770 |
| eco_816 | GSM106755 |
| eco_817 | GSM18277 |
| eco_82 | GSM106337 |
| eco_822 | GSM99154 |
| eco_828 | GSM65596 |
| eco_837 | E-MEXP-267-raw-data-495080287.txt |
| eco_838 | GSM114615 |
| eco_841 | GSM37076 |
| eco_845 | GSM118604 |
| eco_850 | GSM88918 |
| eco_851 | GSM99112 |
| eco_855 | GSM99161 |
| eco_865 | Clean_DIAUX6 |
| eco_89 | GSM111420 |
| eco_9 | 5287 |
| eco_90 | GSM111421 |
| eco_94 | GSM114614 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_2170_1 | 0.2500 | 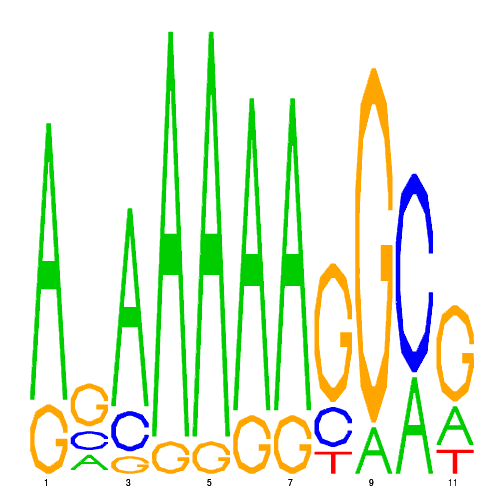 |
GREs assigned to eco_2170
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
Corems associated with eco_2170
| Corem |
|---|