eco_25360 Co-expression
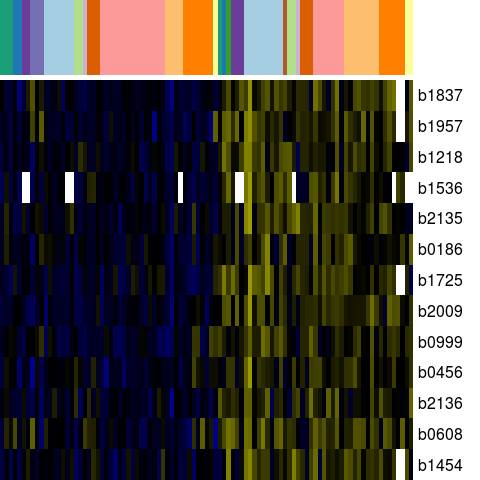
Genes in eco_25360
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0186 | ldcC | NP_414728.1 | lysine decarboxylase 2, constitutive | 209679 | 211820 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0456 | ybaA | NP_414989.1 | hypothetical protein | 475896 | 476249 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0608 | ybdR | NP_415141.1 | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | 641311 | 642549 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0999 | cbpM | NP_415519.1 | modulator of CbpA co-chaperone | 1062078 | 1061773 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1218 | chaC | NP_415736.2 | regulatory protein for cation transpor | 1271730 | 1272425 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1454 | yncG | NP_415971.1 | predicted enzyme | 1524271 | 1524888 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1536 | ydeI | NP_416054.1 | hypothetical protein | 1622521 | 1622129 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1725 | yniA | NP_416239.1 | predicted phosphotransferase/kinase | 1805820 | 1806680 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1837 | yebW | NP_416351.2 | orf, hypothetical protein | 1920145 | 1920336 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1957 | yodC | NP_416466.1 | hypothetical protein | 2026394 | 2026212 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2009 | sbmC | NP_416513.1 | DNA gyrase inhibitor | 2079286 | 2078813 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
Conditions in eco_25360
| Condition ID | Condition Name |
|---|---|
| eco_1 | 1282 |
| eco_103 | GSM120166 |
| eco_122 | GSM18273 |
| eco_123 | GSM18279 |
| eco_132 | GSM37071 |
| eco_133 | GSM37075 |
| eco_151 | GSM73124 |
| eco_161 | GSM73151 |
| eco_169 | GSM73258 |
| eco_180 | GSM73288 |
| eco_183 | GSM77351 |
| eco_185 | GSM77354 |
| eco_19 | Clean_DIAUX7 |
| eco_199 | GSM83058 |
| eco_2 | 1285 |
| eco_202 | GSM83065 |
| eco_221 | GSM99108 |
| eco_242 | GSM99186 |
| eco_247 | GSM99203 |
| eco_249 | GSM99211 |
| eco_256 | GSM99401 |
| eco_257 | GSM99637 |
| eco_263 | 1639 |
| eco_27 | Clean_TGC18 |
| eco_274 | Clean_H2O26 |
| eco_285 | EC_LAC_LA_20_C |
| eco_322 | GSM118613 |
| eco_325 | GSM118621 |
| eco_327 | GSM1367 |
| eco_328 | GSM1372 |
| eco_351 | GSM73122 |
| eco_363 | GSM77358 |
| eco_38 | EC_GLY_GLYA_44_C |
| eco_39 | EC_GLY_GLYB_44_A |
| eco_400 | GSM99157 |
| eco_408 | GSM99195 |
| eco_41 | EC_GLY_GLYC_20_C |
| eco_416 | 1290 |
| eco_418 | 14831 |
| eco_430 | EC_GLY_GLY1_20_A |
| eco_432 | EC_GLY_GLYC_44_C |
| eco_435 | EC_LAC_LC_20_C |
| eco_452 | GSM114372 |
| eco_46 | EC_LAC_L2_60_C |
| eco_462 | GSM23459 |
| eco_465 | GSM37068 |
| eco_476 | GSM65628 |
| eco_480 | GSM67659 |
| eco_492 | GSM77348 |
| eco_494 | GSM77355 |
| eco_496 | GSM82600 |
| eco_5 | 1913 |
| eco_502 | GSM98720 |
| eco_522 | GSM99213 |
| eco_528 | wt__G |
| eco_539 | crpcysB__G |
| eco_582 | GSM63891 |
| eco_591 | GSM82608 |
| eco_596 | GSM83076 |
| eco_597 | GSM83082 |
| eco_607 | GSM99141 |
| eco_642 | E-MEXP-267-raw-data-495080335.txt |
| eco_651 | GSM18236 |
| eco_653 | GSM18274 |
| eco_66 | E-MEXP-245-raw-data-396455381.txt |
| eco_660 | GSM37070 |
| eco_67 | E-MEXP-267-raw-data-495080207.txt |
| eco_679 | GSM8112 |
| eco_682 | GSM83054 |
| eco_701 | GSM99190 |
| eco_705 | GSM99397 |
| eco_713 | EC_LAC_LD_20_C |
| eco_715 | E-MEXP-245-raw-data-396455397.txt |
| eco_732 | GSM73133 |
| eco_74 | GSM101233 |
| eco_755 | Clean_DIAUX2 |
| eco_768 | GSM18278 |
| eco_770 | GSM37073 |
| eco_773 | GSM73153 |
| eco_784 | crp__G |
| eco_787 | fruR__G |
| eco_788 | GSM101238 |
| eco_789 | GSM101239 |
| eco_798 | GSM73253 |
| eco_80 | GSM101772 |
| eco_801 | GSM82547 |
| eco_807 | GSM99636 |
| eco_823 | EC_GLY_GLYB_44_B |
| eco_824 | E-MEXP-222-raw-data-386285069.txt |
| eco_841 | GSM37076 |
| eco_844 | GSM99214 |
| eco_850 | GSM88918 |
| eco_857 | EC_LAC_LC_60_C |
| eco_9 | 5287 |
| eco_99 | GSM118607 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_25360_1 | 0.8200 | 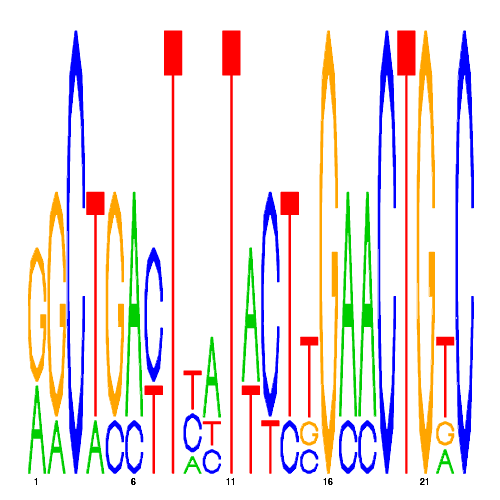 |
| eco_25360_2 | 11.0000 | 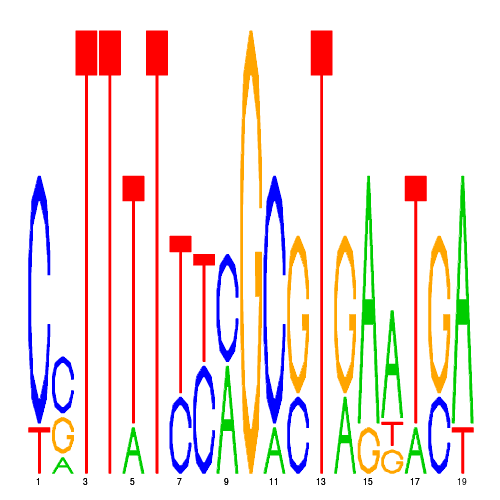 |
GREs assigned to eco_25360
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_65 |  |
Corems associated with eco_25360
| Corem |
|---|
| ec518257 |
| ec522655 |
| ec528619 |
| ec575299 |
| ec578825 |