eco_29950 Co-expression
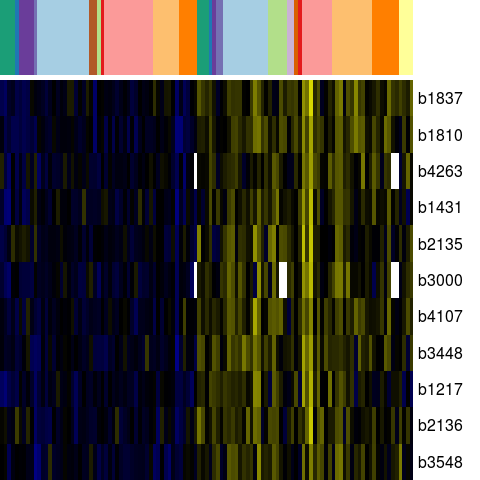
Genes in eco_29950
Conditions in eco_29950
| Condition ID | Condition Name |
|---|---|
| eco_108 | GSM1373 |
| eco_111 | GSM18235 |
| eco_119 | GSM18262 |
| eco_123 | GSM18279 |
| eco_124 | GSM18285 |
| eco_125 | GSM30815 |
| eco_129 | GSM35417 |
| eco_130 | GSM37067 |
| eco_135 | GSM37886 |
| eco_142 | GSM63892 |
| eco_152 | GSM73125 |
| eco_163 | GSM73155 |
| eco_168 | GSM73257 |
| eco_17 | Clean_DIAUX13 |
| eco_187 | GSM7886 |
| eco_19 | Clean_DIAUX7 |
| eco_197 | GSM82616 |
| eco_199 | GSM83058 |
| eco_207 | GSM83080 |
| eco_223 | GSM99113 |
| eco_233 | GSM99150 |
| eco_245 | GSM99196 |
| eco_274 | Clean_H2O26 |
| eco_279 | EC_GLY_GLY2_20_C |
| eco_283 | EC_GLY_GLYE_44_B |
| eco_308 | GSM101777 |
| eco_311 | GSM106340 |
| eco_319 | GSM116025 |
| eco_321 | GSM118611 |
| eco_325 | GSM118621 |
| eco_334 | GSM18266 |
| eco_354 | GSM73147 |
| eco_367 | GSM8117 |
| eco_369 | GSM82543 |
| eco_373 | GSM82598 |
| eco_374 | GSM82604 |
| eco_379 | GSM83073 |
| eco_382 | GSM83083 |
| eco_394 | GSM99125 |
| eco_404 | GSM99183 |
| eco_405 | GSM99187 |
| eco_409 | GSM99200 |
| eco_411 | GSM99208 |
| eco_413 | GSM99391 |
| eco_417 | 14829 |
| eco_43 | EC_GLY_WILD_D |
| eco_434 | EC_LAC_LA_60_B |
| eco_444 | GSM101767 |
| eco_445 | GSM101768 |
| eco_453 | GSM114385 |
| eco_46 | EC_LAC_L2_60_C |
| eco_470 | GSM38548 |
| eco_474 | GSM65594 |
| eco_48 | EC_LAC_LA_60_A |
| eco_481 | GSM73119 |
| eco_482 | GSM73123 |
| eco_486 | GSM73261 |
| eco_495 | GSM8111 |
| eco_496 | GSM82600 |
| eco_498 | GSM82610 |
| eco_505 | GSM99083 |
| eco_507 | GSM99088 |
| eco_512 | GSM99131 |
| eco_518 | GSM99188 |
| eco_523 | GSM99389 |
| eco_527 | wt |
| eco_529 | 1292 |
| eco_536 | Clean_H2O25 |
| eco_539 | crpcysB__G |
| eco_540 | crpfruR__G |
| eco_543 | EC_GLY_GLY1_20_B |
| eco_544 | EC_GLY_GLY2_44_A |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_548 | EC_GLY_GLYE_44_C |
| eco_562 | GADXW_exp12 |
| eco_564 | GSM101780 |
| eco_575 | GSM18253 |
| eco_58 | EC_LAC_WILD_F |
| eco_585 | GSM73132 |
| eco_595 | GSM83070 |
| eco_6 | 1914 |
| eco_60 | E-MEXP-222-raw-data-386284973.txt |
| eco_614 | GSM99210 |
| eco_619 | 1909 |
| eco_636 | EC_LAC_L3_60_B |
| eco_657 | GSM30818 |
| eco_659 | GSM37066 |
| eco_669 | GSM67665 |
| eco_684 | GSM83067 |
| eco_69 | fur__G |
| eco_708 | Clean_ANA9 |
| eco_715 | E-MEXP-245-raw-data-396455397.txt |
| eco_716 | GADXW_exp2 |
| eco_733 | GSM73142 |
| eco_736 | GSM77346 |
| eco_750 | GSM99178 |
| eco_754 | Clean_ANA17 |
| eco_757 | EC_GLY_GLYC_44_B |
| eco_760 | EC_LAC_LD_60_B |
| eco_769 | GSM18282 |
| eco_790 | GSM101774 |
| eco_795 | GSM18246 |
| eco_802 | GSM82620 |
| eco_829 | GSM67663 |
| eco_83 | GSM106341 |
| eco_84 | GSM106757 |
| eco_844 | GSM99214 |
| eco_845 | GSM118604 |
| eco_851 | GSM99112 |
| eco_862 | GSM99386 |
| eco_863 | GSM73118 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_29950_2 | 0.6700 | 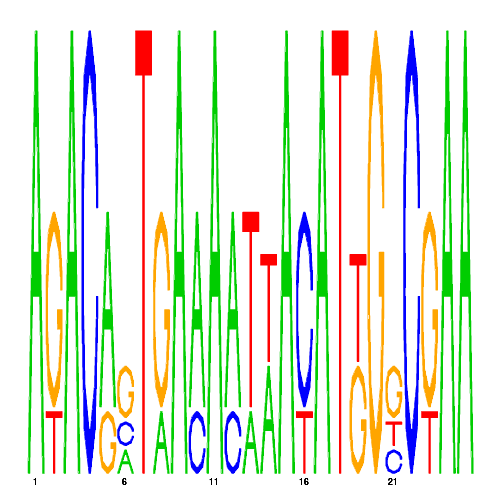 |
| eco_29950_3 | 100.0000 | 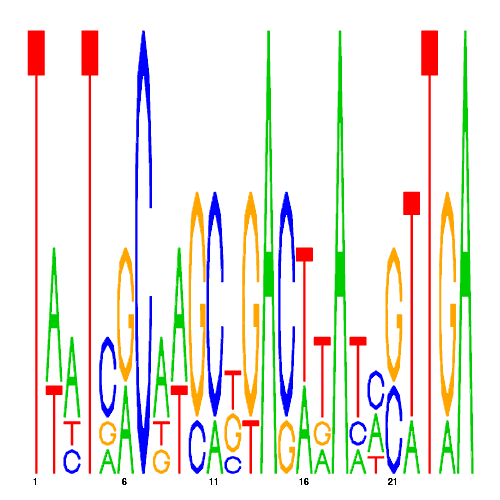 |
| eco_29950_1 | 0.0680 | 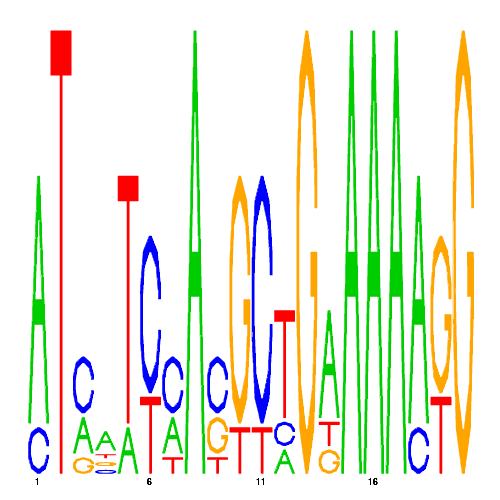 |
GREs assigned to eco_29950
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_0 |  |
| eco_65 |  |
Corems associated with eco_29950
| Corem |
|---|
| ec522655 |
| ec522658 |
| ec522662 |
| ec524226 |
| ec575306 |