eco_35150 Co-expression
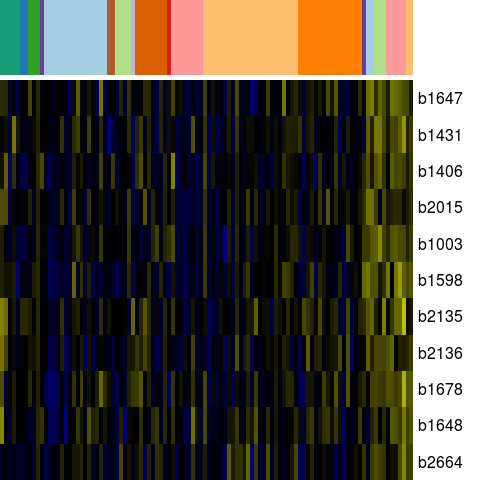
Genes in eco_35150
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1003 | yccJ | NP_415523.1 | hypothetical protein | 1066314 | 1066087 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1406 | ydbC | NP_415924.1 | predicted oxidoreductase, NAD(P)-binding | 1472245 | 1473105 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1431 | ydcL | NP_415948.1 | predicted lipoprotein | 1500481 | 1501149 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1598 | ydgD | NP_416115.1 | predicted peptidase | 1669984 | 1670805 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1647 | ydhF | YP_025305.1 | predicted oxidoreductase | 1723656 | 1722760 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1648 | b1648 | NP_416165.2 | orf, hypothetical protein | 1723944 | 1723705 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1678 | ynhG | NP_416193.1 | hypothetical protein | 1756749 | 1755745 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2015 | yeeY | NP_416519.4 | putative transcriptional regulator LYSR-type | 2086282 | 2085353 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2664 | ygaE | NP_417150.4 | putative transcriptional regulator | 2793696 | 2794358 | + | NC_000913.2 |
Conditions in eco_35150
| Condition ID | Condition Name |
|---|---|
| eco_10 | 8455 |
| eco_107 | GSM137283 |
| eco_116 | GSM18256 |
| eco_124 | GSM18285 |
| eco_137 | GSM62194 |
| eco_149 | GSM67647 |
| eco_156 | GSM73140 |
| eco_171 | GSM73264 |
| eco_173 | GSM73270 |
| eco_177 | GSM73277 |
| eco_184 | GSM77353 |
| eco_197 | GSM82616 |
| eco_203 | GSM83069 |
| eco_205 | GSM83077 |
| eco_212 | GSM98723 |
| eco_219 | GSM99103 |
| eco_223 | GSM99113 |
| eco_226 | GSM99121 |
| eco_23 | Clean_H2O24 |
| eco_254 | GSM99395 |
| eco_279 | EC_GLY_GLY2_20_C |
| eco_293 | E-MEXP-244-raw-data-396455098.txt |
| eco_310 | GSM106339 |
| eco_338 | GSM18283 |
| eco_343 | GSM38549 |
| eco_347 | GSM65624 |
| eco_352 | GSM73134 |
| eco_355 | GSM73255 |
| eco_359 | GSM73269 |
| eco_397 | GSM99145 |
| eco_405 | GSM99187 |
| eco_409 | GSM99200 |
| eco_412 | GSM99390 |
| eco_414 | GSM99398 |
| eco_421 | 5266 |
| eco_434 | EC_LAC_LA_60_B |
| eco_444 | GSM101767 |
| eco_447 | GSM106342 |
| eco_457 | GSM1377 |
| eco_467 | GSM37074 |
| eco_482 | GSM73123 |
| eco_486 | GSM73261 |
| eco_49 | EC_LAC_LA_60_D |
| eco_491 | GSM77344 |
| eco_492 | GSM77348 |
| eco_494 | GSM77355 |
| eco_496 | GSM82600 |
| eco_505 | GSM99083 |
| eco_509 | GSM99099 |
| eco_512 | GSM99131 |
| eco_521 | GSM99199 |
| eco_524 | GSM99394 |
| eco_539 | crpcysB__G |
| eco_575 | GSM18253 |
| eco_582 | GSM63891 |
| eco_586 | GSM73149 |
| eco_589 | GSM77359 |
| eco_590 | GSM77361 |
| eco_608 | GSM99151 |
| eco_612 | GSM99206 |
| eco_616 | 1278 |
| eco_620 | Clean_DIAUX14 |
| eco_646 | GSM116027 |
| eco_656 | GSM18289 |
| eco_657 | GSM30818 |
| eco_665 | GSM65622 |
| eco_670 | GSM73126 |
| eco_671 | GSM73131 |
| eco_672 | GSM73148 |
| eco_677 | GSM73280 |
| eco_685 | GSM83068 |
| eco_687 | GSM88917 |
| eco_69 | fur__G |
| eco_690 | GSM99082 |
| eco_692 | GSM99097 |
| eco_710 | crpfruR |
| eco_713 | EC_LAC_LD_20_C |
| eco_721 | GSM18265 |
| eco_732 | GSM73133 |
| eco_734 | GSM73252 |
| eco_754 | Clean_ANA17 |
| eco_757 | EC_GLY_GLYC_44_B |
| eco_761 | EC_LAC_WILD_A |
| eco_766 | GSM116031 |
| eco_769 | GSM18282 |
| eco_77 | GSM101240 |
| eco_777 | GSM83059 |
| eco_779 | GSM99127 |
| eco_78 | GSM101244 |
| eco_782 | GSM99209 |
| eco_79 | GSM101771 |
| eco_797 | GSM73150 |
| eco_80 | GSM101772 |
| eco_806 | GSM99175 |
| eco_809 | 1287 |
| eco_814 | EC_GLY_WILD_A |
| eco_817 | GSM18277 |
| eco_819 | GSM77352 |
| eco_835 | EC_GLY_GLYB_20_B |
| eco_840 | GSM18272 |
| eco_841 | GSM37076 |
| eco_846 | GSM18248 |
| eco_850 | GSM88918 |
| eco_851 | GSM99112 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_35150_1 | 3.3000 | 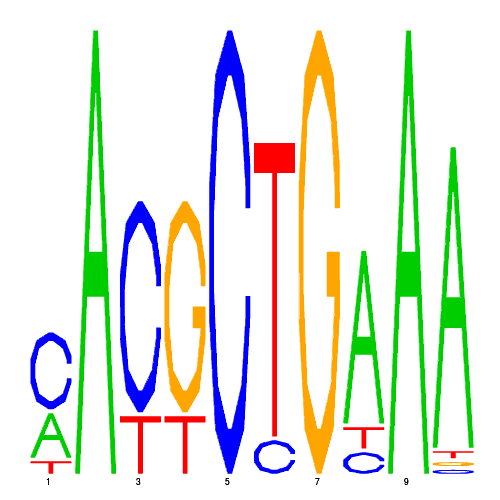 |
GREs assigned to eco_35150
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
Corems associated with eco_35150
| Corem |
|---|