eco_38647 Co-expression
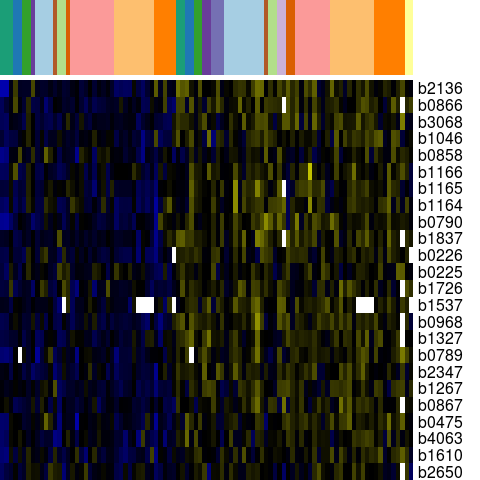
Genes in eco_38647
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0225 | yafQ | NP_414760.1 | predicted toxin of the YafQ-DinJ toxin-antitoxin system | 246239 | 245961 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0226 | dinJ | NP_414761.1 | predicted antitoxin of YafQ-DinJ toxin-antitoxin system | 246502 | 246242 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0475 | hemH | NP_415008.1 | ferrochelatase | 497279 | 498241 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0789 | ybhO | NP_415310.1 | cardiolipin synthase 2 | 822962 | 821721 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0790 | ybhP | NP_415311.1 | predicted DNase | 823720 | 822959 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0858 | ybjO | NP_415379.1 | predicted inner membrane protein | 897212 | 897700 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0866 | ybjQ | NP_415387.1 | hypothetical protein | 903816 | 904139 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0867 | ybjR | NP_415388.1 | predicted amidase and lipoprotein | 904136 | 904966 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0968 | yccX | NP_415488.1 | predicted acylphosphatase | 1029287 | 1029565 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1046 | ymdC | NP_415564.2 | putative synthase | 1105578 | 1106999 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1164 | ycgZ | NP_415682.1 | hypothetical protein | 1215012 | 1215248 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1165 | ymgA | NP_415683.1 | hypothetical protein | 1215291 | 1215563 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1166 | ymgB | NP_415684.1 | hypothetical protein | 1215592 | 1215858 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1267 | yciO | NP_415783.6 | orf, hypothetical protein | 1322122 | 1322742 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1327 | ycjY | NP_415843.2 | predicted hydrolase | 1389877 | 1388957 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1537 | ydeJ | NP_416055.1 | competence damage-inducible protein A | 1622797 | 1623315 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1610 | tus | NP_416127.1 | DNA replication terminus site-binding protein | 1682283 | 1683212 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1726 | yniB | NP_416240.1 | predicted inner membrane protein | 1807257 | 1806721 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1837 | yebW | NP_416351.2 | orf, hypothetical protein | 1920145 | 1920336 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2347 | yfdC | NP_416849.1 | predicted inner membrane protein | 2463323 | 2464255 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2650 | ypjC | hypothetical protein | 2783033 | 2781660 | - | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b3068 | ygjF | NP_417540.1 | G/U mismatch-specific DNA glycosylase | 3213495 | 3212989 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b4063 | soxR | NP_418487.1 | DNA-binding transcriptional dual regulator, Fe-S center for redox-sensing | 4275492 | 4275956 | + | NC_000913.2 |
Conditions in eco_38647
| Condition ID | Condition Name |
|---|---|
| eco_104 | GSM120192 |
| eco_115 | GSM18255 |
| eco_118 | GSM18260 |
| eco_133 | GSM37075 |
| eco_163 | GSM73155 |
| eco_164 | GSM73157 |
| eco_169 | GSM73258 |
| eco_173 | GSM73270 |
| eco_175 | GSM73272 |
| eco_177 | GSM73277 |
| eco_178 | GSM73283 |
| eco_195 | GSM82603 |
| eco_199 | GSM83058 |
| eco_200 | GSM83060 |
| eco_228 | GSM99128 |
| eco_230 | GSM99132 |
| eco_248 | GSM99205 |
| eco_256 | GSM99401 |
| eco_268 | 5284 |
| eco_280 | EC_GLY_GLYA_20_B |
| eco_284 | EC_LAC_LA_20_B |
| eco_287 | EC_LAC_LB_20_C |
| eco_29 | crpfur |
| eco_291 | E-MEXP-222-raw-data-386285007.txt |
| eco_298 | fur |
| eco_326 | GSM118623 |
| eco_330 | GSM1378 |
| eco_360 | GSM73273 |
| eco_369 | GSM82543 |
| eco_370 | GSM82550 |
| eco_373 | GSM82598 |
| eco_395 | GSM99138 |
| eco_400 | GSM99157 |
| eco_401 | GSM99162 |
| eco_41 | EC_GLY_GLYC_20_C |
| eco_412 | GSM99390 |
| eco_417 | 14829 |
| eco_429 | crpcysB |
| eco_437 | E-MEXP-267-raw-data-495080271.txt |
| eco_438 | fruR |
| eco_444 | GSM101767 |
| eco_452 | GSM114372 |
| eco_457 | GSM1377 |
| eco_461 | GSM18284 |
| eco_472 | GSM63896 |
| eco_480 | GSM67659 |
| eco_495 | GSM8111 |
| eco_5 | 1913 |
| eco_500 | GSM88913 |
| eco_501 | GSM88915 |
| eco_512 | GSM99131 |
| eco_513 | GSM99134 |
| eco_523 | GSM99389 |
| eco_528 | wt__G |
| eco_532 | 1915 |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_546 | EC_GLY_GLYD_44_B |
| eco_547 | EC_GLY_GLYE_20_B |
| eco_550 | EC_LAC_L2_60_A |
| eco_556 | EC_LAC_LD_60_C |
| eco_557 | E-MEXP-245-raw-data-396455349.txt |
| eco_573 | GSM137282 |
| eco_578 | GSM18288 |
| eco_583 | GSM63894 |
| eco_588 | GSM77349 |
| eco_589 | GSM77359 |
| eco_595 | GSM83070 |
| eco_604 | GSM99123 |
| eco_643 | GSM101766 |
| eco_645 | GSM106338 |
| eco_648 | GSM118606 |
| eco_65 | E-MEXP-245-raw-data-396455365.txt |
| eco_670 | GSM73126 |
| eco_677 | GSM73280 |
| eco_684 | GSM83067 |
| eco_688 | GSM89487 |
| eco_69 | fur__G |
| eco_691 | GSM99094 |
| eco_696 | GSM99116 |
| eco_744 | GSM99085 |
| eco_762 | GADXW_exp10 |
| eco_763 | GADXW_exp5 |
| eco_767 | GSM18254 |
| eco_787 | fruR__G |
| eco_793 | GSM116029 |
| eco_794 | GSM118601 |
| eco_80 | GSM101772 |
| eco_803 | GSM98724 |
| eco_808 | GSM99638 |
| eco_814 | EC_GLY_WILD_A |
| eco_816 | GSM106755 |
| eco_839 | GSM1370 |
| eco_84 | GSM106757 |
| eco_843 | GSM99126 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_38647_1 | 17.0000 | 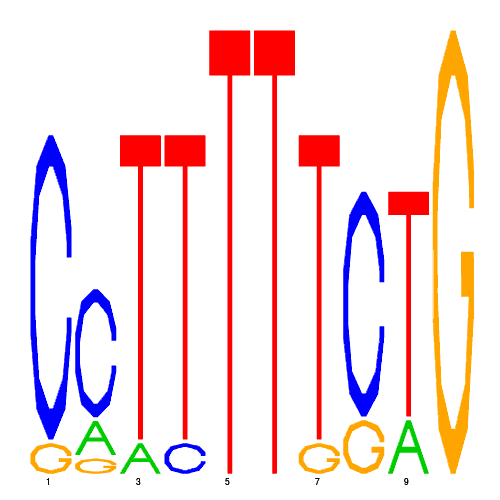 |
| eco_38647_2 | 56000.0000 | 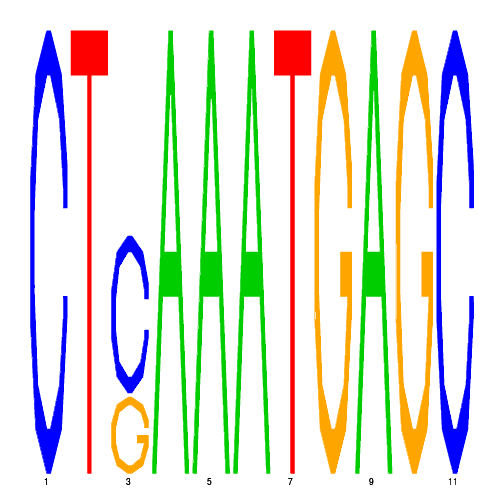 |
GREs assigned to eco_38647
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
| eco_0 |  |
Corems associated with eco_38647
| Corem |
|---|
| ec522655 |
| ec522658 |
| ec613265 |