eco_39746 Co-expression
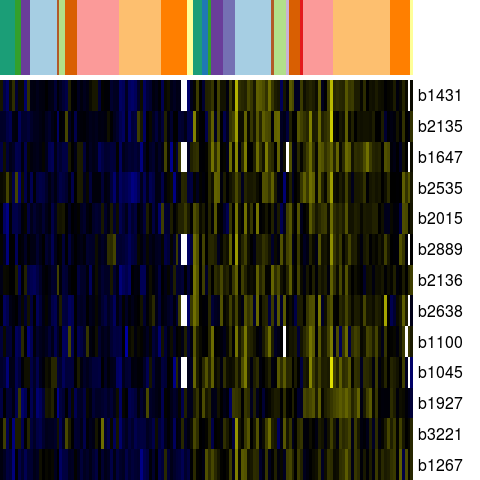
Genes in eco_39746
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1045 | ymdB | NP_415563.1 | hypothetical protein | 1105043 | 1105576 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1100 | ycfH | NP_415618.1 | predicted metallodependent hydrolase | 1156000 | 1156797 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1267 | yciO | NP_415783.6 | orf, hypothetical protein | 1322122 | 1322742 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1431 | ydcL | NP_415948.1 | predicted lipoprotein | 1500481 | 1501149 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1647 | ydhF | YP_025305.1 | predicted oxidoreductase | 1723656 | 1722760 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1927 | amyA | NP_416437.1 | cytoplasmic alpha-amylase | 2004180 | 2005667 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2015 | yeeY | NP_416519.4 | putative transcriptional regulator LYSR-type | 2086282 | 2085353 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2535 | csiE | NP_417030.2 | orf, hypothetical protein | 2663457 | 2664737 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2638 | yfjU | CP4-57 prophage; conserved protein | 2770176 | 2770024 | - | NC_000913.2 | |
| Escherichia coli K-12 MG1655 | b2889 | idi | NP_417365.1 | isopentenyl-diphosphate delta-isomerase | 3031087 | 3031635 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b3221 | yhcH | NP_417688.1 | hypothetical protein | 3367500 | 3367036 | - | NC_000913.2 |
Conditions in eco_39746
| Condition ID | Condition Name |
|---|---|
| eco_104 | GSM120192 |
| eco_107 | GSM137283 |
| eco_115 | GSM18255 |
| eco_124 | GSM18285 |
| eco_126 | GSM30817 |
| eco_132 | GSM37071 |
| eco_133 | GSM37075 |
| eco_135 | GSM37886 |
| eco_140 | GSM63887 |
| eco_153 | GSM73130 |
| eco_16 | Clean_DIAUX12 |
| eco_161 | GSM73151 |
| eco_167 | GSM73160 |
| eco_169 | GSM73258 |
| eco_171 | GSM73264 |
| eco_175 | GSM73272 |
| eco_177 | GSM73277 |
| eco_181 | GSM77341 |
| eco_189 | GSM8119 |
| eco_203 | GSM83069 |
| eco_206 | GSM83078 |
| eco_214 | GSM98727 |
| eco_218 | GSM99096 |
| eco_22 | Clean_H2O22 |
| eco_221 | GSM99108 |
| eco_226 | GSM99121 |
| eco_228 | GSM99128 |
| eco_230 | GSM99132 |
| eco_231 | GSM99133 |
| eco_235 | GSM99159 |
| eco_247 | GSM99203 |
| eco_249 | GSM99211 |
| eco_250 | GSM99215 |
| eco_256 | GSM99401 |
| eco_258 | 1277 |
| eco_267 | 5278 |
| eco_273 | Clean_H2O23 |
| eco_284 | EC_LAC_LA_20_B |
| eco_285 | EC_LAC_LA_20_C |
| eco_289 | EC_LAC_LE_20_B |
| eco_296 | E-MEXP-267-raw-data-495080351.txt |
| eco_301 | GADXW_exp9 |
| eco_309 | GSM101779 |
| eco_32 | EC_GLY_GLY1_44_A |
| eco_347 | GSM65624 |
| eco_359 | GSM73269 |
| eco_36 | EC_GLY_GLYA_20_C |
| eco_361 | GSM73281 |
| eco_364 | GSM77360 |
| eco_365 | GSM8113 |
| eco_379 | GSM83073 |
| eco_383 | GSM88912 |
| eco_400 | GSM99157 |
| eco_404 | GSM99183 |
| eco_407 | GSM99193 |
| eco_415 | GSM99400 |
| eco_418 | 14831 |
| eco_43 | EC_GLY_WILD_D |
| eco_435 | EC_LAC_LC_20_C |
| eco_44 | EC_LAC_L2_20_C |
| eco_442 | GADXW_exp15 |
| eco_444 | GSM101767 |
| eco_456 | GSM118622 |
| eco_459 | GSM18237 |
| eco_46 | EC_LAC_L2_60_C |
| eco_472 | GSM63896 |
| eco_481 | GSM73119 |
| eco_486 | GSM73261 |
| eco_489 | GSM73282 |
| eco_491 | GSM77344 |
| eco_493 | GSM77350 |
| eco_496 | GSM82600 |
| eco_505 | GSM99083 |
| eco_506 | GSM99086 |
| eco_510 | GSM99102 |
| eco_513 | GSM99134 |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_547 | EC_GLY_GLYE_20_B |
| eco_550 | EC_LAC_L2_60_A |
| eco_553 | EC_LAC_LC_20_B |
| eco_561 | E-MEXP-267-raw-data-495080319.txt |
| eco_568 | GSM118602 |
| eco_57 | EC_LAC_WILD_E |
| eco_574 | GSM1375 |
| eco_577 | GSM18287 |
| eco_578 | GSM18288 |
| eco_583 | GSM63894 |
| eco_585 | GSM73132 |
| eco_593 | GSM83061 |
| eco_6 | 1914 |
| eco_600 | GSM98719 |
| eco_604 | GSM99123 |
| eco_606 | GSM99137 |
| eco_626 | EC_GLY_GLY2_20_B |
| eco_629 | EC_GLY_GLYC_20_A |
| eco_634 | EC_LAC_L2_20_A |
| eco_636 | EC_LAC_L3_60_B |
| eco_64 | E-MEXP-245-raw-data-396455333.txt |
| eco_643 | GSM101766 |
| eco_644 | GSM101776 |
| eco_653 | GSM18274 |
| eco_675 | GSM73274 |
| eco_679 | GSM8112 |
| eco_683 | GSM83063 |
| eco_685 | GSM83068 |
| eco_686 | GSM83072 |
| eco_689 | GSM98729 |
| eco_69 | fur__G |
| eco_691 | GSM99094 |
| eco_705 | GSM99397 |
| eco_706 | 1912 |
| eco_721 | GSM18265 |
| eco_734 | GSM73252 |
| eco_740 | GSM89485 |
| eco_755 | Clean_DIAUX2 |
| eco_758 | EC_GLY_GLYD_44_C |
| eco_760 | EC_LAC_LD_60_B |
| eco_767 | GSM18254 |
| eco_775 | GSM73289 |
| eco_783 | Clean_DIAUX8 |
| eco_794 | GSM118601 |
| eco_798 | GSM73253 |
| eco_80 | GSM101772 |
| eco_803 | GSM98724 |
| eco_81 | GSM101773 |
| eco_817 | GSM18277 |
| eco_818 | GSM73129 |
| eco_837 | E-MEXP-267-raw-data-495080287.txt |
| eco_841 | GSM37076 |
| eco_842 | GSM65639 |
| eco_846 | GSM18248 |
| eco_853 | GSM73128 |
| eco_855 | GSM99161 |
| eco_856 | Clean_ANA20 |
| eco_858 | GSM118609 |
| eco_859 | GSM18280 |
| eco_861 | GSM99124 |
| eco_95 | GSM116024 |
| eco_96 | GSM116026 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_39746_2 | 28.0000 | 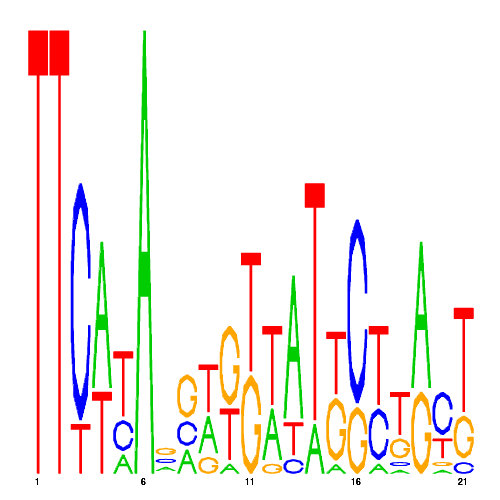 |
| eco_39746_1 | 2.2000 | 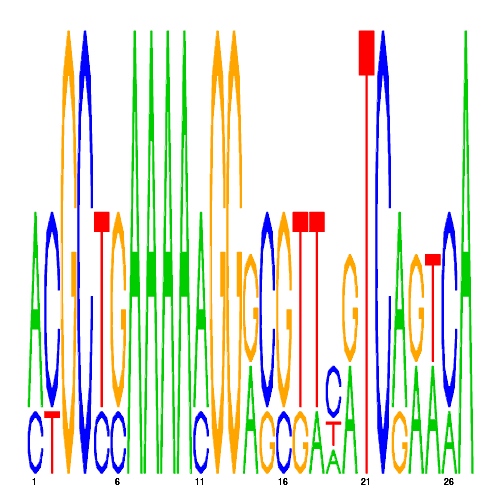 |
GREs assigned to eco_39746
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_65 |  |
Corems associated with eco_39746
| Corem |
|---|
| ec522658 |
| ec613265 |