eco_40490 Co-expression
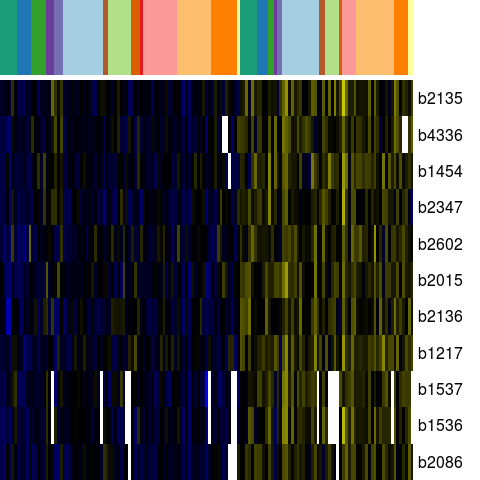
Genes in eco_40490
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1217 | chaB | NP_415735.1 | cation transport regulator | 1271342 | 1271572 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1454 | yncG | NP_415971.1 | predicted enzyme | 1524271 | 1524888 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1536 | ydeI | NP_416054.1 | hypothetical protein | 1622521 | 1622129 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1537 | ydeJ | NP_416055.1 | competence damage-inducible protein A | 1622797 | 1623315 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2015 | yeeY | NP_416519.4 | putative transcriptional regulator LYSR-type | 2086282 | 2085353 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2086 | yegS | NP_416590.1 | hypothetical protein | 2166736 | 2167635 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2347 | yfdC | NP_416849.1 | predicted inner membrane protein | 2463323 | 2464255 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2602 | yfiL | NP_417093.2 | orf, hypothetical protein | 2739382 | 2739747 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b4336 | yjiN | NP_418756.1 | conserved inner membrane protein | 4565269 | 4563989 | - | NC_000913.2 |
Conditions in eco_40490
| Condition ID | Condition Name |
|---|---|
| eco_10 | 8455 |
| eco_100 | GSM118612 |
| eco_106 | GSM1371 |
| eco_112 | GSM18247 |
| eco_116 | GSM18256 |
| eco_118 | GSM18260 |
| eco_124 | GSM18285 |
| eco_127 | GSM30820 |
| eco_138 | GSM62195 |
| eco_143 | GSM63893 |
| eco_155 | GSM73139 |
| eco_156 | GSM73140 |
| eco_157 | GSM73141 |
| eco_167 | GSM73160 |
| eco_169 | GSM73258 |
| eco_172 | GSM73265 |
| eco_195 | GSM82603 |
| eco_2 | 1285 |
| eco_200 | GSM83060 |
| eco_205 | GSM83077 |
| eco_216 | GSM99090 |
| eco_221 | GSM99108 |
| eco_229 | GSM99129 |
| eco_231 | GSM99133 |
| eco_232 | GSM99136 |
| eco_253 | GSM99393 |
| eco_256 | GSM99401 |
| eco_261 | 1595 |
| eco_262 | 1638 |
| eco_268 | 5284 |
| eco_270 | Clean_ANA3 |
| eco_274 | Clean_H2O26 |
| eco_276 | cysB__G |
| eco_298 | fur |
| eco_305 | GSM101241 |
| eco_310 | GSM106339 |
| eco_320 | GSM116030 |
| eco_322 | GSM118613 |
| eco_336 | GSM18275 |
| eco_337 | GSM18281 |
| eco_338 | GSM18283 |
| eco_344 | GSM62196 |
| eco_355 | GSM73255 |
| eco_357 | GSM73262 |
| eco_361 | GSM73281 |
| eco_364 | GSM77360 |
| eco_379 | GSM83073 |
| eco_383 | GSM88912 |
| eco_409 | GSM99200 |
| eco_416 | 1290 |
| eco_419 | 1637 |
| eco_42 | EC_GLY_GLYD_20_C |
| eco_421 | 5266 |
| eco_423 | 9395 |
| eco_424 | Clean_ANA2 |
| eco_429 | crpcysB |
| eco_43 | EC_GLY_WILD_D |
| eco_439 | GADXW_exp1 |
| eco_44 | EC_LAC_L2_20_C |
| eco_446 | GSM101769 |
| eco_449 | GSM111414 |
| eco_45 | EC_LAC_L2_60_B |
| eco_460 | GSM18259 |
| eco_470 | GSM38548 |
| eco_481 | GSM73119 |
| eco_483 | GSM73136 |
| eco_489 | GSM73282 |
| eco_49 | EC_LAC_LA_60_D |
| eco_496 | GSM82600 |
| eco_500 | GSM88913 |
| eco_501 | GSM88915 |
| eco_511 | GSM99104 |
| eco_514 | GSM99135 |
| eco_519 | GSM99194 |
| eco_524 | GSM99394 |
| eco_538 | crp |
| eco_540 | crpfruR__G |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_560 | E-MEXP-267-raw-data-495080303.txt |
| eco_567 | GSM111423 |
| eco_570 | GSM118618 |
| eco_573 | GSM137282 |
| eco_575 | GSM18253 |
| eco_58 | EC_LAC_WILD_F |
| eco_580 | GSM37887 |
| eco_592 | GSM83056 |
| eco_600 | GSM98719 |
| eco_604 | GSM99123 |
| eco_608 | GSM99151 |
| eco_610 | GSM99174 |
| eco_617 | 1641 |
| eco_621 | Clean_DIAUX15 |
| eco_622 | Clean_DIAUX16 |
| eco_623 | Clean_H2O21 |
| eco_626 | EC_GLY_GLY2_20_B |
| eco_631 | EC_GLY_GLYE_44_A |
| eco_641 | E-MEXP-244-raw-data-396455050.txt |
| eco_643 | GSM101766 |
| eco_644 | GSM101776 |
| eco_645 | GSM106338 |
| eco_656 | GSM18289 |
| eco_662 | GSM63890 |
| eco_676 | GSM73275 |
| eco_679 | GSM8112 |
| eco_680 | GSM8114 |
| eco_681 | GSM82546 |
| eco_682 | GSM83054 |
| eco_683 | GSM83063 |
| eco_687 | GSM88917 |
| eco_690 | GSM99082 |
| eco_701 | GSM99190 |
| eco_705 | GSM99397 |
| eco_707 | Clean_ANA15 |
| eco_708 | Clean_ANA9 |
| eco_726 | GSM63888 |
| eco_736 | GSM77346 |
| eco_740 | GSM89485 |
| eco_746 | GSM99110 |
| eco_75 | GSM101234 |
| eco_760 | EC_LAC_LD_60_B |
| eco_762 | GADXW_exp10 |
| eco_764 | GSM101243 |
| eco_77 | GSM101240 |
| eco_771 | GSM73127 |
| eco_776 | GSM77342 |
| eco_78 | GSM101244 |
| eco_783 | Clean_DIAUX8 |
| eco_787 | fruR__G |
| eco_79 | GSM101771 |
| eco_791 | GSM114383 |
| eco_793 | GSM116029 |
| eco_794 | GSM118601 |
| eco_795 | GSM18246 |
| eco_80 | GSM101772 |
| eco_817 | GSM18277 |
| eco_818 | GSM73129 |
| eco_82 | GSM106337 |
| eco_826 | GSM106756 |
| eco_833 | GSM99177 |
| eco_841 | GSM37076 |
| eco_845 | GSM118604 |
| eco_850 | GSM88918 |
| eco_851 | GSM99112 |
| eco_88 | GSM111417 |
| eco_92 | GSM114368 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_40490_2 | 0.5800 | 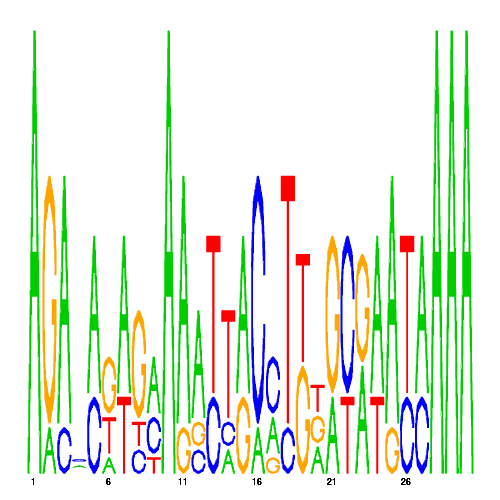 |
| eco_40490_1 | 0.3500 | 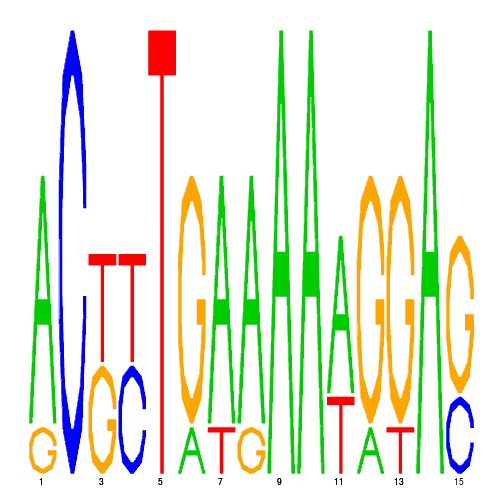 |
GREs assigned to eco_40490
| GRE ID | PSSM |
|---|---|
| eco_0 |  |
| eco_65 |  |
Corems associated with eco_40490
| Corem |
|---|
| ec522655 |
| ec522658 |
| ec613265 |