eco_40733 Co-expression
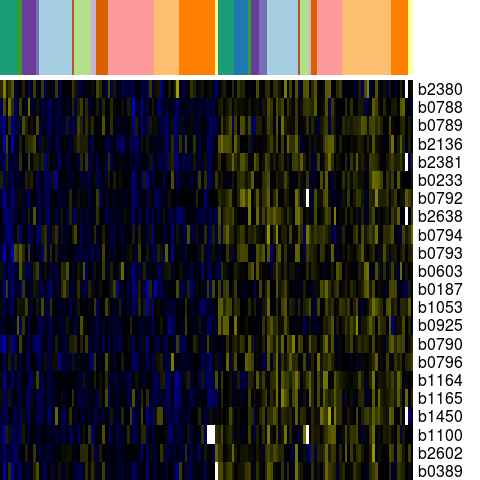
Genes in eco_40733
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0187 | yaeR | NP_414729.4 | orf, hypothetical protein | 211877 | 212266 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0233 | yafO | NP_414768.1 | predicted toxin of the YafO-YafN toxin-antitoxin system | 252301 | 252699 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0389 | yaiA | NP_414923.1 | hypothetical protein | 406203 | 406394 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0603 | ybdO | NP_415136.1 | predicted DNA-binding transcriptional regulator | 636841 | 635939 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0788 | ybhN | NP_415309.1 | conserved inner membrane protein | 821721 | 820765 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0789 | ybhO | NP_415310.1 | cardiolipin synthase 2 | 822962 | 821721 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0790 | ybhP | NP_415311.1 | predicted DNase | 823720 | 822959 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0792 | ybhR | NP_415313.1 | predicted transporter subunit: membrane component of ABC superfamily | 825331 | 824225 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0793 | ybhS | NP_415314.1 | predicted transporter subunit: membrane component of ABC superfamily | 826475 | 825342 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0794 | ybhF | NP_415315.4 | putative ATP-binding component of a transport system | 828204 | 826468 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0796 | ybiH | NP_415317.4 | putative transcriptional regulator | 829866 | 829195 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0925 | ycbB | NP_415445.1 | predicted carboxypeptidase | 980270 | 982117 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1053 | mdtG | NP_415571.1 | predicted drug efflux system | 1114713 | 1113487 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1100 | ycfH | NP_415618.1 | predicted metallodependent hydrolase | 1156000 | 1156797 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1164 | ycgZ | NP_415682.1 | hypothetical protein | 1215012 | 1215248 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1165 | ymgA | NP_415683.1 | hypothetical protein | 1215291 | 1215563 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1450 | b1450 | NP_415967.2 | orf, hypothetical protein | 1518286 | 1518951 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2380 | ypdA | NP_416881.1 | predicted sensory kinase in two-component system with Ypd | 2496693 | 2498390 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2381 | ypdB | NP_416882.1 | predicted response regulator in two-component system withYpdA | 2498405 | 2499139 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2602 | yfiL | NP_417093.2 | orf, hypothetical protein | 2739382 | 2739747 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2638 | yfjU | CP4-57 prophage; conserved protein | 2770176 | 2770024 | - | NC_000913.2 |
Conditions in eco_40733
| Condition ID | Condition Name |
|---|---|
| eco_10 | 8455 |
| eco_100 | GSM118612 |
| eco_102 | GSM118620 |
| eco_103 | GSM120166 |
| eco_118 | GSM18260 |
| eco_124 | GSM18285 |
| eco_138 | GSM62195 |
| eco_141 | GSM63889 |
| eco_143 | GSM63893 |
| eco_144 | GSM65590 |
| eco_147 | GSM65630 |
| eco_157 | GSM73141 |
| eco_160 | GSM73146 |
| eco_167 | GSM73160 |
| eco_169 | GSM73258 |
| eco_172 | GSM73265 |
| eco_185 | GSM77354 |
| eco_195 | GSM82603 |
| eco_196 | GSM82613 |
| eco_2 | 1285 |
| eco_200 | GSM83060 |
| eco_205 | GSM83077 |
| eco_229 | GSM99129 |
| eco_231 | GSM99133 |
| eco_235 | GSM99159 |
| eco_238 | GSM99167 |
| eco_245 | GSM99196 |
| eco_249 | GSM99211 |
| eco_254 | GSM99395 |
| eco_256 | GSM99401 |
| eco_268 | 5284 |
| eco_271 | Clean_DIAUX4 |
| eco_274 | Clean_H2O26 |
| eco_275 | Clean_H2O28 |
| eco_276 | cysB__G |
| eco_281 | EC_GLY_GLYD_20_A |
| eco_302 | GSM101231 |
| eco_305 | GSM101241 |
| eco_31 | EC_GLY_GLY1_20_C |
| eco_331 | GSM1381 |
| eco_336 | GSM18275 |
| eco_337 | GSM18281 |
| eco_338 | GSM18283 |
| eco_343 | GSM38549 |
| eco_344 | GSM62196 |
| eco_355 | GSM73255 |
| eco_357 | GSM73262 |
| eco_361 | GSM73281 |
| eco_374 | GSM82604 |
| eco_379 | GSM83073 |
| eco_38 | EC_GLY_GLYA_44_C |
| eco_381 | GSM83081 |
| eco_385 | GSM98718 |
| eco_391 | GSM99105 |
| eco_395 | GSM99138 |
| eco_40 | EC_GLY_GLYC_20_B |
| eco_409 | GSM99200 |
| eco_416 | 1290 |
| eco_421 | 5266 |
| eco_423 | 9395 |
| eco_424 | Clean_ANA2 |
| eco_429 | crpcysB |
| eco_430 | EC_GLY_GLY1_20_A |
| eco_435 | EC_LAC_LC_20_C |
| eco_436 | EC_LAC_LE_20_A |
| eco_439 | GADXW_exp1 |
| eco_44 | EC_LAC_L2_20_C |
| eco_445 | GSM101768 |
| eco_446 | GSM101769 |
| eco_45 | EC_LAC_L2_60_B |
| eco_468 | GSM37885 |
| eco_470 | GSM38548 |
| eco_481 | GSM73119 |
| eco_483 | GSM73136 |
| eco_489 | GSM73282 |
| eco_49 | EC_LAC_LA_60_D |
| eco_494 | GSM77355 |
| eco_496 | GSM82600 |
| eco_500 | GSM88913 |
| eco_504 | GSM98728 |
| eco_507 | GSM99088 |
| eco_513 | GSM99134 |
| eco_514 | GSM99135 |
| eco_516 | GSM99169 |
| eco_519 | GSM99194 |
| eco_525 | HEATSHOCK_CH |
| eco_538 | crp |
| eco_540 | crpfruR__G |
| eco_542 | EC_GLU_WILD_B |
| eco_545 | EC_GLY_GLYD_44_A |
| eco_548 | EC_GLY_GLYE_44_C |
| eco_549 | EC_LAC_L2_20_B |
| eco_551 | EC_LAC_LA_60_C |
| eco_552 | EC_LAC_LB_60_A |
| eco_56 | EC_LAC_LE_60_C |
| eco_560 | E-MEXP-267-raw-data-495080303.txt |
| eco_567 | GSM111423 |
| eco_570 | GSM118618 |
| eco_575 | GSM18253 |
| eco_58 | EC_LAC_WILD_F |
| eco_580 | GSM37887 |
| eco_59 | E-MEXP-222-raw-data-386284924.txt |
| eco_592 | GSM83056 |
| eco_600 | GSM98719 |
| eco_604 | GSM99123 |
| eco_607 | GSM99141 |
| eco_608 | GSM99151 |
| eco_620 | Clean_DIAUX14 |
| eco_621 | Clean_DIAUX15 |
| eco_622 | Clean_DIAUX16 |
| eco_623 | Clean_H2O21 |
| eco_631 | EC_GLY_GLYE_44_A |
| eco_638 | EC_LAC_LB_60_B |
| eco_643 | GSM101766 |
| eco_647 | GSM116028 |
| eco_656 | GSM18289 |
| eco_66 | E-MEXP-245-raw-data-396455381.txt |
| eco_662 | GSM63890 |
| eco_667 | GSM67657 |
| eco_669 | GSM67665 |
| eco_679 | GSM8112 |
| eco_683 | GSM83063 |
| eco_691 | GSM99094 |
| eco_7 | 5268 |
| eco_700 | GSM99179 |
| eco_705 | GSM99397 |
| eco_708 | Clean_ANA9 |
| eco_717 | GADXW_exp4 |
| eco_726 | GSM63888 |
| eco_736 | GSM77346 |
| eco_740 | GSM89485 |
| eco_75 | GSM101234 |
| eco_752 | GSM99182 |
| eco_762 | GADXW_exp10 |
| eco_766 | GSM116031 |
| eco_77 | GSM101240 |
| eco_771 | GSM73127 |
| eco_772 | GSM73143 |
| eco_783 | Clean_DIAUX8 |
| eco_787 | fruR__G |
| eco_79 | GSM101771 |
| eco_794 | GSM118601 |
| eco_795 | GSM18246 |
| eco_796 | GSM63886 |
| eco_80 | GSM101772 |
| eco_817 | GSM18277 |
| eco_841 | GSM37076 |
| eco_850 | GSM88918 |
| eco_852 | GSM99156 |
| eco_87 | GSM111415 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_40733_1 | 0.0220 | 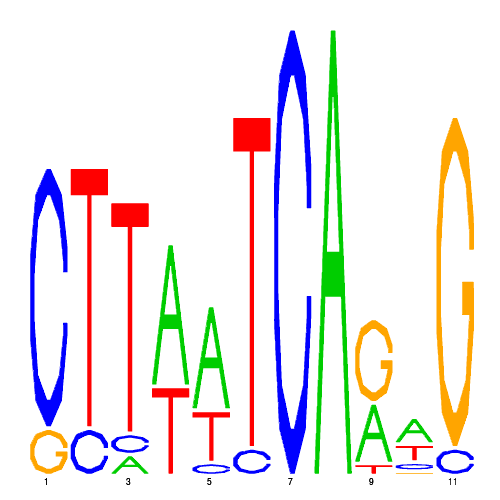 |
| eco_40733_2 | 3600.0000 | 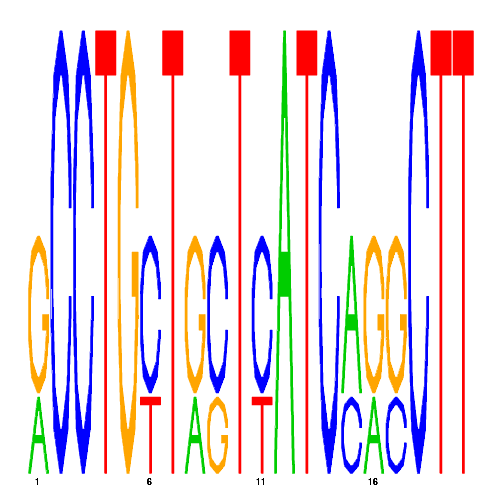 |
GREs assigned to eco_40733
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
| eco_0 |  |
Corems associated with eco_40733
| Corem |
|---|
| ec519441 |
| ec529316 |
| ec613265 |