eco_41561 Co-expression
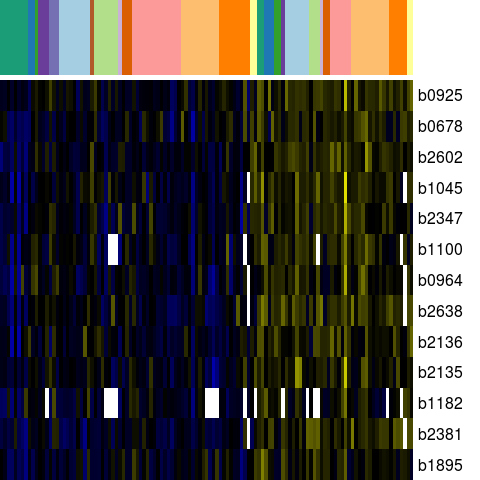
Genes in eco_41561
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b0678 | nagB | NP_415204.1 | glucosamine-6-phosphate deaminase | 702834 | 702034 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0925 | ycbB | NP_415445.1 | predicted carboxypeptidase | 980270 | 982117 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b0964 | yccT | NP_415484.1 | hypothetical protein | 1026996 | 1026334 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1045 | ymdB | NP_415563.1 | hypothetical protein | 1105043 | 1105576 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1100 | ycfH | NP_415618.1 | predicted metallodependent hydrolase | 1156000 | 1156797 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1182 | hlyE | NP_415700.4 | hemolysin E | 1229617 | 1228706 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1895 | yecG | NP_416409.1 | universal stress protein | 1977777 | 1978205 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2136 | yohD | NP_416640.2 | orf, hypothetical protein | 2223823 | 2224401 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2347 | yfdC | NP_416849.1 | predicted inner membrane protein | 2463323 | 2464255 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2381 | ypdB | NP_416882.1 | predicted response regulator in two-component system withYpdA | 2498405 | 2499139 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2602 | yfiL | NP_417093.2 | orf, hypothetical protein | 2739382 | 2739747 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2638 | yfjU | CP4-57 prophage; conserved protein | 2770176 | 2770024 | - | NC_000913.2 |
Conditions in eco_41561
| Condition ID | Condition Name |
|---|---|
| eco_1 | 1282 |
| eco_100 | GSM118612 |
| eco_107 | GSM137283 |
| eco_112 | GSM18247 |
| eco_115 | GSM18255 |
| eco_119 | GSM18262 |
| eco_12 | Clean_ANA13 |
| eco_133 | GSM37075 |
| eco_156 | GSM73140 |
| eco_157 | GSM73141 |
| eco_166 | GSM73159 |
| eco_167 | GSM73160 |
| eco_168 | GSM73257 |
| eco_173 | GSM73270 |
| eco_187 | GSM7886 |
| eco_197 | GSM82616 |
| eco_201 | GSM83062 |
| eco_21 | Clean_H2O211 |
| eco_219 | GSM99103 |
| eco_223 | GSM99113 |
| eco_239 | GSM99173 |
| eco_247 | GSM99203 |
| eco_273 | Clean_H2O23 |
| eco_28 | Clean_TGC2 |
| eco_296 | E-MEXP-267-raw-data-495080351.txt |
| eco_298 | fur |
| eco_309 | GSM101779 |
| eco_31 | EC_GLY_GLY1_20_C |
| eco_333 | GSM18258 |
| eco_336 | GSM18275 |
| eco_347 | GSM65624 |
| eco_349 | GSM67648 |
| eco_359 | GSM73269 |
| eco_390 | GSM99101 |
| eco_394 | GSM99125 |
| eco_396 | GSM99143 |
| eco_401 | GSM99162 |
| eco_405 | GSM99187 |
| eco_412 | GSM99390 |
| eco_416 | 1290 |
| eco_417 | 14829 |
| eco_420 | 1908 |
| eco_435 | EC_LAC_LC_20_C |
| eco_439 | GADXW_exp1 |
| eco_446 | GSM101769 |
| eco_452 | GSM114372 |
| eco_454 | GSM118600 |
| eco_46 | EC_LAC_L2_60_C |
| eco_481 | GSM73119 |
| eco_483 | GSM73136 |
| eco_484 | GSM73154 |
| eco_501 | GSM88915 |
| eco_506 | GSM99086 |
| eco_507 | GSM99088 |
| eco_514 | GSM99135 |
| eco_516 | GSM99169 |
| eco_524 | GSM99394 |
| eco_525 | HEATSHOCK_CH |
| eco_526 | STATIONARYPHASE_CH |
| eco_530 | 14830 |
| eco_536 | Clean_H2O25 |
| eco_539 | crpcysB__G |
| eco_546 | EC_GLY_GLYD_44_B |
| eco_547 | EC_GLY_GLYE_20_B |
| eco_55 | EC_LAC_LE_60_A |
| eco_56 | EC_LAC_LE_60_C |
| eco_574 | GSM1375 |
| eco_575 | GSM18253 |
| eco_588 | GSM77349 |
| eco_59 | E-MEXP-222-raw-data-386284924.txt |
| eco_600 | GSM98719 |
| eco_610 | GSM99174 |
| eco_619 | 1909 |
| eco_621 | Clean_DIAUX15 |
| eco_622 | Clean_DIAUX16 |
| eco_626 | EC_GLY_GLY2_20_B |
| eco_628 | EC_GLY_GLYB_20_A |
| eco_634 | EC_LAC_L2_20_A |
| eco_642 | E-MEXP-267-raw-data-495080335.txt |
| eco_644 | GSM101776 |
| eco_651 | GSM18236 |
| eco_656 | GSM18289 |
| eco_66 | E-MEXP-245-raw-data-396455381.txt |
| eco_660 | GSM37070 |
| eco_667 | GSM67657 |
| eco_671 | GSM73131 |
| eco_678 | GSM77356 |
| eco_680 | GSM8114 |
| eco_688 | GSM89487 |
| eco_69 | fur__G |
| eco_70 | GADXW_exp13 |
| eco_704 | GSM99380 |
| eco_710 | crpfruR |
| eco_717 | GADXW_exp4 |
| eco_748 | GSM99163 |
| eco_750 | GSM99178 |
| eco_752 | GSM99182 |
| eco_754 | Clean_ANA17 |
| eco_767 | GSM18254 |
| eco_776 | GSM77342 |
| eco_784 | crp__G |
| eco_786 | EC_LAC_WILD_C |
| eco_787 | fruR__G |
| eco_79 | GSM101771 |
| eco_790 | GSM101774 |
| eco_795 | GSM18246 |
| eco_815 | GSM101770 |
| eco_816 | GSM106755 |
| eco_819 | GSM77352 |
| eco_825 | GADXW_exp3 |
| eco_828 | GSM65596 |
| eco_83 | GSM106341 |
| eco_832 | GSM99168 |
| eco_836 | EC_GLY_GLYD_20_B |
| eco_845 | GSM118604 |
| eco_85 | GSM106758 |
| eco_87 | GSM111415 |
| eco_88 | GSM111417 |
| eco_9 | 5287 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_41561_1 | 0.2600 | 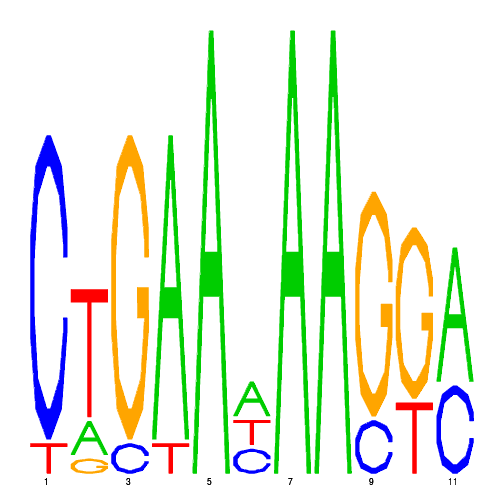 |
GREs assigned to eco_41561
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
Corems associated with eco_41561
| Corem |
|---|
| ec522655 |
| ec522658 |
| ec528631 |
| ec613265 |