eco_42774 Co-expression
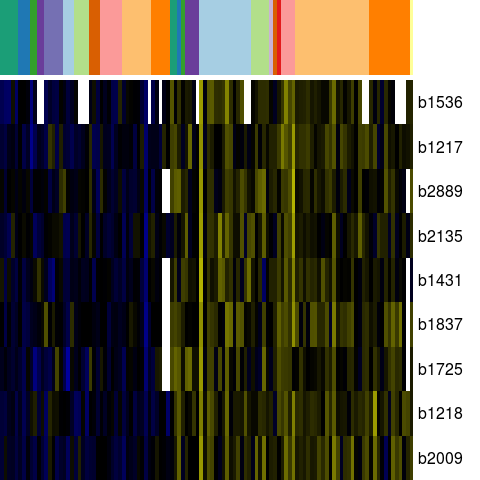
Genes in eco_42774
| Organism | Gene name | Common name | Accession | Description | Start | Stop | Strand | Chromosome |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli K-12 MG1655 | b1217 | chaB | NP_415735.1 | cation transport regulator | 1271342 | 1271572 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1218 | chaC | NP_415736.2 | regulatory protein for cation transpor | 1271730 | 1272425 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1431 | ydcL | NP_415948.1 | predicted lipoprotein | 1500481 | 1501149 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1536 | ydeI | NP_416054.1 | hypothetical protein | 1622521 | 1622129 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1725 | yniA | NP_416239.1 | predicted phosphotransferase/kinase | 1805820 | 1806680 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b1837 | yebW | NP_416351.2 | orf, hypothetical protein | 1920145 | 1920336 | + | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2009 | sbmC | NP_416513.1 | DNA gyrase inhibitor | 2079286 | 2078813 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2135 | yohC | NP_416639.2 | orf, hypothetical protein | 2223653 | 2223066 | - | NC_000913.2 |
| Escherichia coli K-12 MG1655 | b2889 | idi | NP_417365.1 | isopentenyl-diphosphate delta-isomerase | 3031087 | 3031635 | + | NC_000913.2 |
Conditions in eco_42774
| Condition ID | Condition Name |
|---|---|
| eco_109 | GSM1379 |
| eco_123 | GSM18279 |
| eco_126 | GSM30817 |
| eco_130 | GSM37067 |
| eco_131 | GSM37069 |
| eco_132 | GSM37071 |
| eco_152 | GSM73125 |
| eco_153 | GSM73130 |
| eco_158 | GSM73144 |
| eco_163 | GSM73155 |
| eco_169 | GSM73258 |
| eco_172 | GSM73265 |
| eco_173 | GSM73270 |
| eco_176 | GSM73276 |
| eco_193 | GSM82548 |
| eco_195 | GSM82603 |
| eco_201 | GSM83062 |
| eco_213 | GSM98726 |
| eco_220 | GSM99107 |
| eco_227 | GSM99122 |
| eco_230 | GSM99132 |
| eco_24 | Clean_H2O27 |
| eco_247 | GSM99203 |
| eco_250 | GSM99215 |
| eco_254 | GSM99395 |
| eco_256 | GSM99401 |
| eco_268 | 5284 |
| eco_272 | Clean_DIAUX9 |
| eco_28 | Clean_TGC2 |
| eco_29 | crpfur |
| eco_293 | E-MEXP-244-raw-data-396455098.txt |
| eco_294 | E-MEXP-245-raw-data-396455413.txt |
| eco_296 | E-MEXP-267-raw-data-495080351.txt |
| eco_310 | GSM106339 |
| eco_312 | GSM106504 |
| eco_315 | GSM111418 |
| eco_322 | GSM118613 |
| eco_323 | GSM118614 |
| eco_325 | GSM118621 |
| eco_33 | EC_GLY_GLY1_44_B |
| eco_367 | GSM8117 |
| eco_374 | GSM82604 |
| eco_383 | GSM88912 |
| eco_393 | GSM99118 |
| eco_404 | GSM99183 |
| eco_407 | GSM99193 |
| eco_413 | GSM99391 |
| eco_415 | GSM99400 |
| eco_417 | 14829 |
| eco_431 | EC_GLY_GLY2_44_C |
| eco_432 | EC_GLY_GLYC_44_C |
| eco_441 | GADXW_exp14 |
| eco_446 | GSM101769 |
| eco_454 | GSM118600 |
| eco_457 | GSM1377 |
| eco_462 | GSM23459 |
| eco_477 | GSM65634 |
| eco_481 | GSM73119 |
| eco_482 | GSM73123 |
| eco_485 | GSM73156 |
| eco_505 | GSM99083 |
| eco_509 | GSM99099 |
| eco_513 | GSM99134 |
| eco_518 | GSM99188 |
| eco_521 | GSM99199 |
| eco_524 | GSM99394 |
| eco_527 | wt |
| eco_541 | crpfur__G |
| eco_557 | E-MEXP-245-raw-data-396455349.txt |
| eco_560 | E-MEXP-267-raw-data-495080303.txt |
| eco_561 | E-MEXP-267-raw-data-495080319.txt |
| eco_577 | GSM18287 |
| eco_584 | GSM73121 |
| eco_588 | GSM77349 |
| eco_59 | E-MEXP-222-raw-data-386284924.txt |
| eco_590 | GSM77361 |
| eco_591 | GSM82608 |
| eco_592 | GSM83056 |
| eco_607 | GSM99141 |
| eco_622 | Clean_DIAUX16 |
| eco_653 | GSM18274 |
| eco_668 | GSM67658 |
| eco_671 | GSM73131 |
| eco_674 | GSM73267 |
| eco_676 | GSM73275 |
| eco_677 | GSM73280 |
| eco_681 | GSM82546 |
| eco_686 | GSM83072 |
| eco_688 | GSM89487 |
| eco_697 | GSM99144 |
| eco_699 | GSM99171 |
| eco_743 | GSM98722 |
| eco_75 | GSM101234 |
| eco_768 | GSM18278 |
| eco_769 | GSM18282 |
| eco_774 | GSM73254 |
| eco_775 | GSM73289 |
| eco_782 | GSM99209 |
| eco_788 | GSM101238 |
| eco_79 | GSM101771 |
| eco_799 | GSM73259 |
| eco_80 | GSM101772 |
| eco_809 | 1287 |
| eco_81 | GSM101773 |
| eco_811 | Clean_DIAUX17 |
| eco_816 | GSM106755 |
| eco_817 | GSM18277 |
| eco_818 | GSM73129 |
| eco_820 | GSM89488 |
| eco_821 | GSM99146 |
| eco_90 | GSM111421 |
| eco_99 | GSM118607 |
De novo detected cis-regulatory motifs
| CRM ID | eval | PSSM |
|---|---|---|
| eco_42774_1 | 12.0000 | 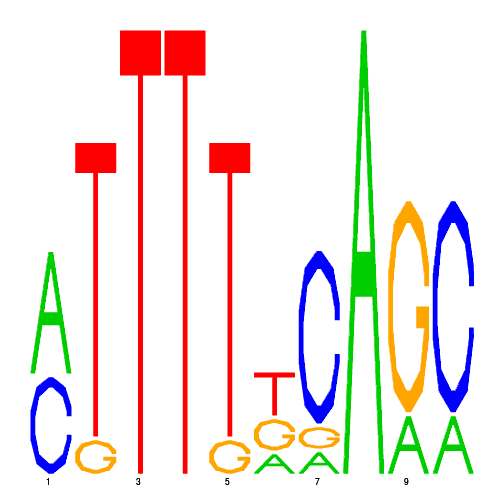 |
GREs assigned to eco_42774
| GRE ID | PSSM |
|---|---|
| eco_65 |  |
Corems associated with eco_42774
| Corem |
|---|
| ec522655 |
| ec522658 |